# A tibble: 7,107 × 19
forested year elevation eastness northness roughness tree_no_tree dew_temp
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <dbl>
1 Yes 2005 881 90 43 63 Tree 0.04
2 Yes 2005 113 -25 96 30 Tree 6.4
3 No 2005 164 -84 53 13 Tree 6.06
4 Yes 2005 299 93 34 6 No tree 4.43
5 Yes 2005 806 47 -88 35 Tree 1.06
6 Yes 2005 736 -27 -96 53 Tree 1.35
7 Yes 2005 636 -48 87 3 No tree 1.42
8 Yes 2005 224 -65 -75 9 Tree 6.39
9 Yes 2005 52 -62 78 42 Tree 6.5
10 Yes 2005 2240 -67 -74 99 No tree -5.63
# ℹ 7,097 more rows
# ℹ 11 more variables: precip_annual <dbl>, temp_annual_mean <dbl>,
# temp_annual_min <dbl>, temp_annual_max <dbl>, temp_january_min <dbl>,
# vapor_min <dbl>, vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>,
# land_type <fct>Data budget/making a model
Lecture 3
Cornell University
INFO 4940/5940 - Fall 2024
September 3, 2024
Announcements
Announcements
- Office hours beginning this week
Learning objectives
- Identify the importance of budgeting data for machine learning
- Partition data into training and test sets
- Specify models using the {parsnip} package
- Utilize workflows to bundle preprocessing and modeling tasks
- Fit models using the {tidymodels} framework
- Generate predictions from the fitted model
Application exercise
ae-02
Install the {renv} package
Go to the course GitHub org and find your
ae-02(repo name will be suffixed with your GitHub name).Clone the repo in RStudio, open the Quarto document in the repo, install the required packages, and follow along and complete the exercises.
Render, commit, and push your edits by the AE deadline – end of the day
Data on forests in Washington
Data on forests in Washington
- The U.S. Forest Service maintains ML models to predict whether a plot of land is “forested.”
- This classification is important for all sorts of research, legislation, and land management purposes.
- Plots are typically remeasured every 10 years and this dataset contains the most recent measurement per plot.
- Type
?forestedto learn more about this dataset, including references.
Data on forests in Washington
N = 7,107plots of land, one from each of 7,107 6000-acre hexagons in WA.- A nominal outcome,
forested, with levels"Yes"and"No", measured “on-the-ground.” - 18 remotely-sensed and easily-accessible predictors:
- numeric variables based on weather and topography.
- nominal variables based on classifications from other governmental orgs.
Checklist for predictors
Is it ethical to use this variable? (Or even legal?)
Will this variable be available at prediction time?
Does this variable contribute to explainability?
Data on forests in Washington
Data splitting and spending
Data splitting and spending
For machine learning, we typically split data into training and test sets:
- The training set is used to estimate model parameters.
- The test set is used to find an independent assessment of model performance.
Do not 🚫 use the test set during training.
Data splitting and spending

The more data we spend
the better estimates we’ll get.
Data splitting and spending
- Spending too much data in training prevents us from computing a good assessment of predictive performance.
- Spending too much data in testing prevents us from computing a good estimate of model parameters.
⏱️ Your turn
When is a good time to split your data?
01:00
The testing data is precious 💎
The initial split
What is set.seed()?
To create that split of the data, R generates “pseudo-random” numbers: while they are made to behave like random numbers, their generation is deterministic given a “seed”.
This allows us to reproduce results by setting that seed.
Which seed you pick doesn’t matter, as long as you don’t try a bunch of seeds and pick the one that gives you the best performance.
Accessing the data
The training set
# A tibble: 5,330 × 19
forested year elevation eastness northness roughness tree_no_tree dew_temp
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <dbl>
1 No 2016 464 -5 -99 7 No tree 1.71
2 Yes 2016 166 92 37 7 Tree 6
3 No 2016 644 -85 -52 24 No tree 0.67
4 Yes 2014 1285 4 99 79 Tree 1.91
5 Yes 2013 822 87 48 68 Tree 1.95
6 Yes 2017 3 6 -99 5 Tree 7.93
7 Yes 2014 2041 -95 28 49 Tree -4.22
8 Yes 2015 1009 -8 99 72 Tree 1.72
9 No 2017 436 -98 19 10 No tree 1.8
10 No 2018 775 63 76 103 No tree 0.62
# ℹ 5,320 more rows
# ℹ 11 more variables: precip_annual <dbl>, temp_annual_mean <dbl>,
# temp_annual_min <dbl>, temp_annual_max <dbl>, temp_january_min <dbl>,
# vapor_min <dbl>, vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>,
# land_type <fct>The test set
🙈️
There are 1777 rows and 19 columns in the test set.
⏱️ Your turn
Split your data so 20% is held out for the test set.
Try out different values in set.seed() to see how the results change.
05:00
Data splitting and spending
Exploratory data analysis for ML 🧐
⏱️ Your turn
Explore the forested_train data on your own!
- What’s the distribution of the outcome,
forested? - What’s the distribution of numeric variables like
precip_annual? - How does the distribution of
foresteddiffer across the categorical variables?
08:00





The whole game - status update

Fitting models in R
Linear models
How do you fit a linear model in R?
lmfor linear modelglmnetfor regularized regressionkerasfor regression using TensorFlowstanfor Bayesian regressionsparkfor large data setsbruleefor regression using torch
To specify a model
- Choose a model
- Specify an engine
- Set the mode
To specify a model
To specify a model
- Choose a model
- Specify an engine
- Set the mode
To specify a model
To specify a model
To specify a model
- Choose a model
- Specify an engine
- Set the mode
To specify a model
To specify a model
Decision Tree Model Specification (classification)
Computational engine: rpart All available models are listed at https://www.tidymodels.org/find/parsnip/
To specify a model
- Choose a model
- Specify an engine
- Set the mode
⏱️ Your turn
Run the tree_spec chunk in your .qmd.
Edit this code to use a logistic regression model.
All available models are listed at https://www.tidymodels.org/find/parsnip/
Extension/Challenge: Edit this code to use a different model. For example, try using a conditional inference tree as implemented in the {partykit} package by changing the engine - or try an entirely different model type!
05:00
Models we’ll use this week
- Logistic regression
- Decision trees
Logistic regression

Logistic regression

Logistic regression

- Logit of outcome probability modeled as linear combination of predictors:
\(\log(\frac{p}{1 - p}) = \beta_0 + \beta_1 \times \text{A}\)
- Find a sigmoid line that separates the two classes
Decision trees

Decision trees

Series of splits or if/then statements based on predictors
First the tree grows until some condition is met (maximum depth, no more data)
Then the tree is pruned to reduce its complexity
Decision trees


All models are wrong, but some are useful!
Logistic regression

Decision trees

A model workflow
Workflows bind preprocessors and models
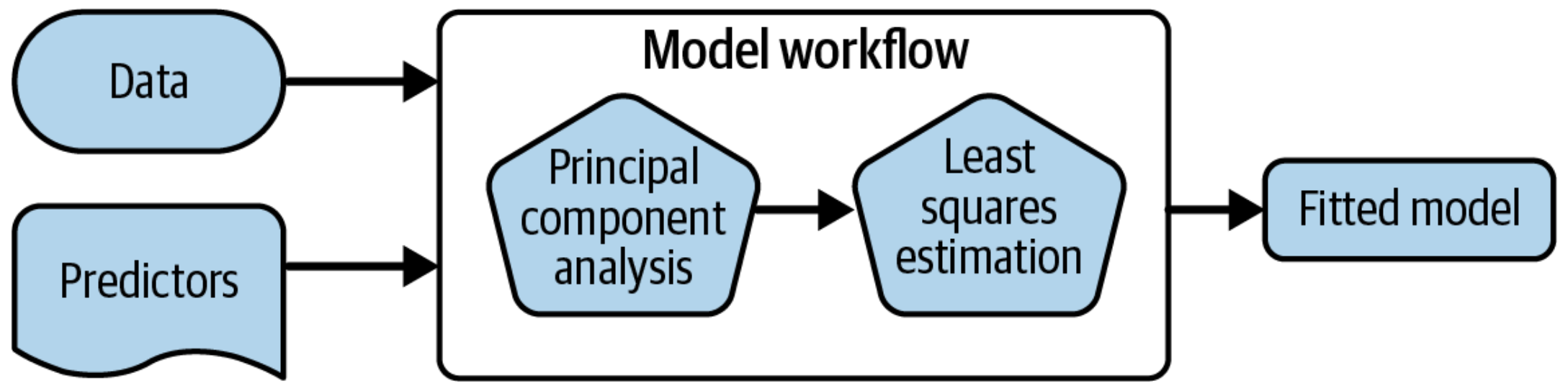
What is wrong with this?
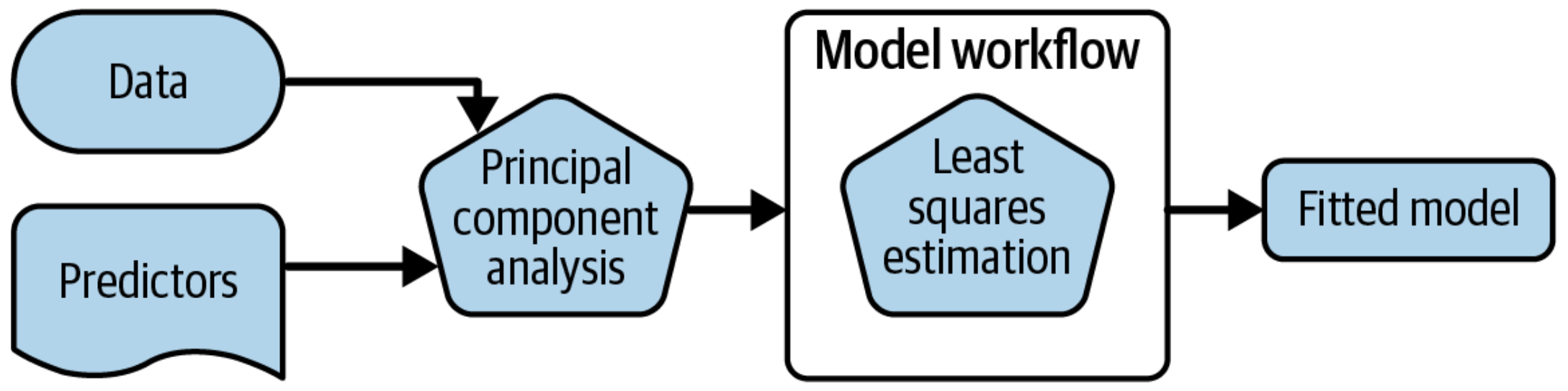
Why a workflow()?
- Workflows handle new data better than base R tools in terms of new factor levels
- You can use other preprocessors besides formulas (see “Feature engineering”)
- They can help organize your work when working with multiple models
- Most importantly, a workflow captures the entire modeling process:
fit()andpredict()apply to the preprocessing steps in addition to the actual model fit
A model workflow
tree_spec <- decision_tree() |>
set_mode("classification")
tree_spec |>
fit(forested ~ ., data = forested_train) parsnip model object
n= 5685
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5685 2550 Yes (0.55145119 0.44854881)
2) land_type=Tree 3064 300 Yes (0.90208877 0.09791123) *
3) land_type=Barren,Non-tree vegetation 2621 371 No (0.14154903 0.85845097)
6) temp_annual_max< 13.395 347 153 Yes (0.55907781 0.44092219)
12) tree_no_tree=Tree 92 6 Yes (0.93478261 0.06521739) *
13) tree_no_tree=No tree 255 108 No (0.42352941 0.57647059) *
7) temp_annual_max>=13.395 2274 177 No (0.07783641 0.92216359) *A model workflow
tree_spec <- decision_tree() |>
set_mode("classification")
workflow() |>
add_formula(forested ~ .) |>
add_model(tree_spec) |>
fit(data = forested_train) ══ Workflow [trained] ════════════════════════════════════════════════
Preprocessor: Formula
Model: decision_tree()
── Preprocessor ──────────────────────────────────────────────────────
forested ~ .
── Model ─────────────────────────────────────────────────────────────
n= 5685
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5685 2550 Yes (0.55145119 0.44854881)
2) land_type=Tree 3064 300 Yes (0.90208877 0.09791123) *
3) land_type=Barren,Non-tree vegetation 2621 371 No (0.14154903 0.85845097)
6) temp_annual_max< 13.395 347 153 Yes (0.55907781 0.44092219)
12) tree_no_tree=Tree 92 6 Yes (0.93478261 0.06521739) *
13) tree_no_tree=No tree 255 108 No (0.42352941 0.57647059) *
7) temp_annual_max>=13.395 2274 177 No (0.07783641 0.92216359) *A model workflow
tree_spec <- decision_tree() |>
set_mode("classification")
workflow(forested ~ ., tree_spec) |>
fit(data = forested_train) ══ Workflow [trained] ════════════════════════════════════════════════
Preprocessor: Formula
Model: decision_tree()
── Preprocessor ──────────────────────────────────────────────────────
forested ~ .
── Model ─────────────────────────────────────────────────────────────
n= 5685
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5685 2550 Yes (0.55145119 0.44854881)
2) land_type=Tree 3064 300 Yes (0.90208877 0.09791123) *
3) land_type=Barren,Non-tree vegetation 2621 371 No (0.14154903 0.85845097)
6) temp_annual_max< 13.395 347 153 Yes (0.55907781 0.44092219)
12) tree_no_tree=Tree 92 6 Yes (0.93478261 0.06521739) *
13) tree_no_tree=No tree 255 108 No (0.42352941 0.57647059) *
7) temp_annual_max>=13.395 2274 177 No (0.07783641 0.92216359) *⏱️ Your turn
Run the tree_wflow chunk in your .qmd.
Edit this code to make a workflow with your own model of choice.
Extension/Challenge: Other than formulas, what kinds of preprocessors are supported?
05:00
Predict with your model
How do you use your new tree_fit model?
⏱️ Your turn
Run:
predict(tree_fit, new_data = forested_test)
What do you notice about the structure of the result?
01:00
⏱️ Your turn
Run:
augment(tree_fit, new_data = forested_test)
How does the output compare to the output from predict()?
01:00
The tidymodels prediction guarantee!
- The predictions will always be inside a tibble
- The column names and types are unsurprising and predictable
- The number of rows in
new_dataand the output are the same
Understand your model
How do you understand your new tree_fit model?

Understand your model
How do you understand your new tree_fit model?
You can extract_*() several components of your fitted workflow.
⚠️ Never predict() with any extracted components!
Understand your model
How do you understand your new tree_fit model?
You can use your fitted workflow for model and/or prediction explanations:
- overall variable importance, such as with the {vip} package
- flexible model explainers, such as with the {DALEXtra} package
Learn more at https://www.tmwr.org/explain
⏱️ Your turn
Extract the model engine object from your fitted workflow and check it out.
02:00
The whole game - status update

Wrap-up
Recap
- Splitting data into training and test sets ensures we have a valid way to assess the final model’s performance
- Exploratory data analysis is important to understanding your data’s structure and (simple) patterns/relationships
- Use {parsnip} to specify machine learning models in R
- Use {workflows} to bundle preprocessing and modeling tasks
Acknowledgments
- Materials derived in part from Machine learning with {tidymodels} and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.
My latest school tax bill
