Tuning hyperparameters
Lecture 7
Cornell University
INFO 4940/5940 - Fall 2024
September 19, 2024
Announcements
Announcements
- No new homework this week
Learning objectives
- Review tuning parameters for machine learning models
- Define boosted trees models
- Utilize grid search to optimize tuning parameters
- Implement space-filling designs for grid searches
- Introduce iterative search procedures for tuning parameters
- Implement Bayesian optimization using the {tune} package
- Finalize the model for production
Application exercise
ae-06
- Go to the course GitHub org and find your
ae-06(repo name will be suffixed with your GitHub name). - Clone the repo in RStudio, open the Quarto document in the repo, install the required packages, and follow along and complete the exercises.
- Render, commit, and push your edits by the AE deadline – end of the day
From last time
Previously - Data Usage
Previously - Feature engineering
library(textrecipes)
hash_rec <- recipe(avg_price_per_room ~ ., data = hotel_train) |>
step_YeoJohnson(lead_time) |>
# Defaults to 32 signed indicator columns
step_dummy_hash(agent) |>
step_dummy_hash(company) |>
# Regular indicators for the others
step_dummy(all_nominal_predictors()) |>
step_zv(all_predictors())Optimizing models via tuning parameters
Tuning parameters
Some model or preprocessing parameters cannot be estimated directly from the data.
Some examples:
- ✅ Tree depth in decision trees
- ✅ Number of neighbors in a K-nearest neighbor model
- ✅ Number of features to hash in a feature hashing recipe
- ❌ Bayesian priors for model parameters
- ❌ The random seed
Optimize tuning parameters
- Try different values and measure their performance.
- Find good values for these parameters.
- Once the value(s) of the parameter(s) are determined, a model can be finalized by fitting the model to the entire training set.
Tagging parameters for tuning
With {tidymodels}, you can mark the parameters that you want to optimize with a value of tune().
The function itself just returns… itself:
Optimizing the hash features
Our new recipe is:
We will be using a tree-based model in a minute.
- The other categorical predictors are left as-is.
- That’s why there is no
step_dummy().
Boosting models
Boosting
- Bagging - use bootstrap resampling to create multiple copies of training data, fit a separate model to each copy, and aggregate the results
- Grow trees independently
- Boosting - grow trees sequentially
How do you study?
Slow, methodical preparation

Cram the night before
Which is more effective in the long run? Learning slowly
Boosted trees
- Fit a decision tree to the residuals from the model
- Add the tree to the model and update the residuals
- Repeat steps 1 and 2 until complete
Each tree is small - only a handful of terminal nodes
Each tree uses the results of the previous tree to better predict samples, especially those that have been poorly predicted.
Each tree in the ensemble is saved and new samples are predicted using a weighted average of the votes of each tree in the ensemble.
Popular implementations
- Adaboost (Adaptive Boosting)
- Gradient boosting
- LightGBM (Light Gradient Boosting Machine)
- XGBoost (Extreme Gradient Boosting)
Boosted tree tuning parameters
Some possible parameters:
mtry: The number of predictors randomly sampled at each split (in \([1, ncol(x)]\) or \((0, 1]\)).trees: The number of trees (\([1, \infty]\), but usually up to thousands)min_n: The number of samples needed to further split (\([1, n]\)).learn_rate: The rate that each tree adapts from previous iterations (\((0, \infty]\), usual maximum is 0.1).stop_iter: The number of iterations of boosting where no improvement was shown before stopping (\([1, trees]\))
Boosted tree tuning parameters
It is usually not difficult to optimize these models.
Often, there are multiple candidate tuning parameter combinations that have very good results.
To demonstrate simple concepts, we’ll look at optimizing the number of trees in the ensemble (between 1 and 100) and the learning rate (\(10^{-5}\) to \(10^{-1}\)).
Boosted tree tuning parameters
We’ll need to load the {bonsai} package. This has the information needed to use LightGBM
Optimize tuning parameters
The main two strategies for optimization are:
Grid search 💠 which tests a pre-defined set of candidate values
Iterative search 🌀 which suggests/estimates new values of candidate parameters to evaluate
Grid search
A small grid of points trying to minimize the error via learning rate:
Grid search
In reality we would probably sample the space more densely:
Iterative Search
We could start with a few points and search the space:
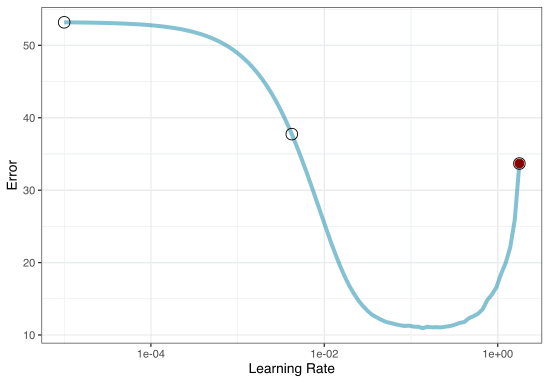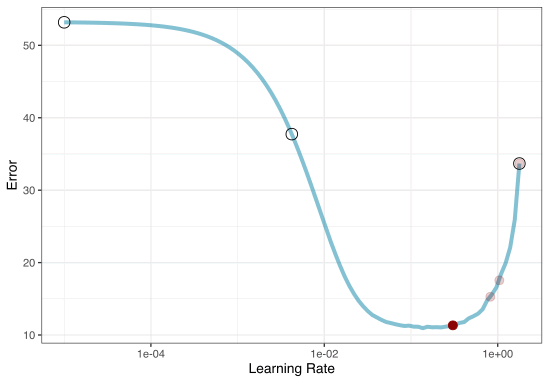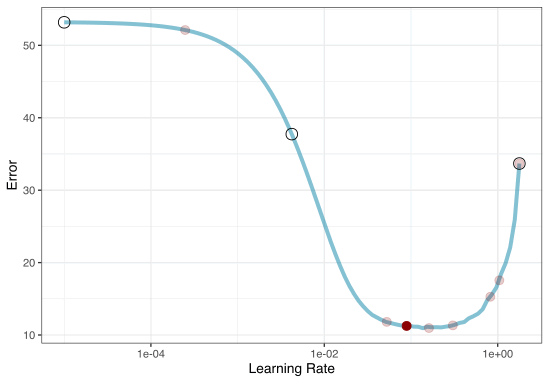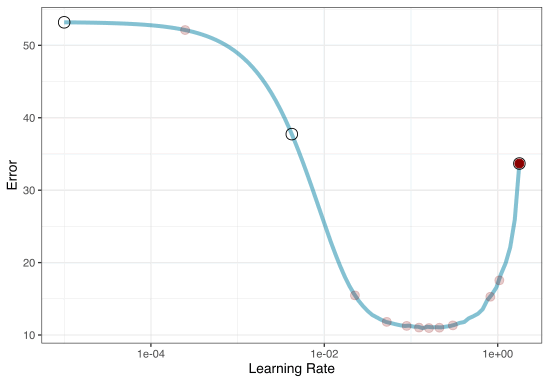
Grid search
Parameters
The {tidymodels} framework provides pre-defined information on tuning parameters (such as their type, range, transformations, etc).
The
extract_parameter_set_dials()function extracts these tuning parameters and the info.
Grids
Create your grid manually or automatically.
The
grid_*()functions can make a grid.
Different types of grids

Space-filling designs (SFD) attempt to cover the parameter space without redundant candidates.
Create a grid
Collection of 4 parameters for tuning
identifier type object
trees trees nparam[+]
learn_rate learn_rate nparam[+]
agent hash num_terms nparam[+]
company hash num_terms nparam[+]# Trees (quantitative)
Range: [1, 2000]Learning Rate (quantitative)
Transformer: log-10 [1e-100, Inf]
Range (transformed scale): [-10, -1]A parameter set can be updated (e.g. to change the ranges).
Create a grid
set.seed(12)
grid <- lgbm_wflow |>
extract_parameter_set_dials() |>
grid_space_filling(size = 25)
grid# A tibble: 25 × 4
trees learn_rate `agent hash` `company hash`
<int> <dbl> <int> <int>
1 1 7.50e- 6 574 574
2 84 1.78e- 5 2048 2298
3 167 5.62e-10 1824 912
4 250 4.22e- 5 3250 512
5 334 1.78e- 8 512 2896
6 417 1.33e- 3 322 1625
7 500 1 e- 1 1448 1149
8 584 1 e- 7 1290 256
9 667 2.37e-10 456 724
10 750 1.78e- 2 645 322
# ℹ 15 more rows⏱️ Your turn
Create a grid for our tunable workflow.
Try creating a regular grid.
03:00
Create a regular grid
# A tibble: 256 × 4
trees learn_rate `agent hash` `company hash`
<int> <dbl> <int> <int>
1 1 0.0000000001 256 256
2 667 0.0000000001 256 256
3 1333 0.0000000001 256 256
4 2000 0.0000000001 256 256
5 1 0.0000001 256 256
6 667 0.0000001 256 256
7 1333 0.0000001 256 256
8 2000 0.0000001 256 256
9 1 0.0001 256 256
10 667 0.0001 256 256
# ℹ 246 more rows⏱️ Your turn
What advantage would a regular grid have?
01:00
Update parameter ranges
lgbm_param <- lgbm_wflow |>
extract_parameter_set_dials() |>
update(
trees = trees(c(1L, 100L)),
learn_rate = learn_rate(c(-5, -1))
)
set.seed(712)
grid <- lgbm_param |>
grid_regular(size = 25)
grid# A tibble: 81 × 4
trees learn_rate `agent hash` `company hash`
<int> <dbl> <int> <int>
1 1 0.00001 256 256
2 50 0.00001 256 256
3 100 0.00001 256 256
4 1 0.001 256 256
5 50 0.001 256 256
6 100 0.001 256 256
7 1 0.1 256 256
8 50 0.1 256 256
9 100 0.1 256 256
10 1 0.00001 1024 256
# ℹ 71 more rowsThe results
Note that the learning rates are uniform on the log-10 scale and this shows 2 of 4 dimensions.
Use the tune_*() functions to tune models
Choosing tuning parameters
Let’s take our previous model and tune more parameters:
lgbm_spec <- boost_tree(trees = tune(), learn_rate = tune(), min_n = tune()) |>
set_mode("regression") |>
set_engine("lightgbm")
lgbm_wflow <- workflow(hash_rec, lgbm_spec)
# Update the feature hash ranges (log-2 units)
lgbm_param <- lgbm_wflow |>
extract_parameter_set_dials() |>
update(
`agent hash` = num_hash(c(3, 8)),
`company hash` = num_hash(c(3, 8))
)Grid search
Grid search
# Tuning results
# 10-fold cross-validation using stratification
# A tibble: 10 × 5
splits id .metrics .notes .predictions
<list> <chr> <list> <list> <list>
1 <split [3372/377]> Fold01 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
2 <split [3373/376]> Fold02 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
3 <split [3373/376]> Fold03 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
4 <split [3373/376]> Fold04 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
5 <split [3373/376]> Fold05 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
6 <split [3374/375]> Fold06 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
7 <split [3375/374]> Fold07 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
8 <split [3376/373]> Fold08 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
9 <split [3376/373]> Fold09 <tibble [50 × 9]> <tibble [0 × 3]> <tibble>
10 <split [3376/373]> Fold10 <tibble [50 × 9]> <tibble [0 × 3]> <tibble> Grid results

Tuning results
# A tibble: 50 × 11
trees min_n learn_rate `agent hash` `company hash` .metric .estimator mean
<int> <int> <dbl> <int> <int> <chr> <chr> <dbl>
1 298 19 4.15e- 9 222 36 mae standard 53.5
2 298 19 4.15e- 9 222 36 rsq standard 0.816
3 1394 5 5.82e- 6 28 21 mae standard 53.2
4 1394 5 5.82e- 6 28 21 rsq standard 0.817
5 774 12 4.41e- 2 27 95 mae standard 9.86
6 774 12 4.41e- 2 27 95 rsq standard 0.949
7 1342 7 6.84e-10 71 17 mae standard 53.5
8 1342 7 6.84e-10 71 17 rsq standard 0.816
9 669 39 8.62e- 7 141 145 mae standard 53.5
10 669 39 8.62e- 7 141 145 rsq standard 0.817
# ℹ 40 more rows
# ℹ 3 more variables: n <int>, std_err <dbl>, .config <chr>Tuning results
# A tibble: 500 × 10
id trees min_n learn_rate `agent hash` `company hash` .metric .estimator
<chr> <int> <int> <dbl> <int> <int> <chr> <chr>
1 Fold01 298 19 4.15e-9 222 36 mae standard
2 Fold01 298 19 4.15e-9 222 36 rsq standard
3 Fold02 298 19 4.15e-9 222 36 mae standard
4 Fold02 298 19 4.15e-9 222 36 rsq standard
5 Fold03 298 19 4.15e-9 222 36 mae standard
6 Fold03 298 19 4.15e-9 222 36 rsq standard
7 Fold04 298 19 4.15e-9 222 36 mae standard
8 Fold04 298 19 4.15e-9 222 36 rsq standard
9 Fold05 298 19 4.15e-9 222 36 mae standard
10 Fold05 298 19 4.15e-9 222 36 rsq standard
# ℹ 490 more rows
# ℹ 2 more variables: .estimate <dbl>, .config <chr>Choose a parameter combination
# A tibble: 5 × 11
trees min_n learn_rate `agent hash` `company hash` .metric .estimator mean
<int> <int> <dbl> <int> <int> <chr> <chr> <dbl>
1 1890 10 0.0159 115 174 rsq standard 0.950
2 774 12 0.0441 27 95 rsq standard 0.949
3 1638 36 0.0409 15 120 rsq standard 0.948
4 963 23 0.00556 157 13 rsq standard 0.937
5 590 5 0.00320 85 73 rsq standard 0.911
# ℹ 3 more variables: n <int>, std_err <dbl>, .config <chr>Choose a parameter combination
Create your own tibble for final parameters or use one of the tune::select_*() functions:
Checking calibration
Running in parallel
Grid search, combined with resampling, requires fitting a lot of models!
These models don’t depend on one another and can be run in parallel.
We can use a parallel backend to do this:

Running in parallel
Speed-ups are fairly linear up to the number of physical cores (10 here).

Early stopping for boosted trees
We have directly optimized the number of trees as a tuning parameter.
Instead we could
- Set the number of trees to a single large number.
- Stop adding trees when performance gets worse.
This is known as “early stopping” and there is a parameter for that: stop_iter.
Early stopping has a potential to decrease the tuning time.
⏱️ Your turn
Set trees = 2000 and tune the stop_iter parameter.
Note that you will need to regenerate lgbm_param with your new workflow!
10:00
lgbm_spec <- boost_tree(trees = 2000, learn_rate = tune(),
min_n = tune(), stop_iter = tune()) |>
set_mode("regression") |>
set_engine("lightgbm")
lgbm_wflow <- workflow(hash_rec, lgbm_spec)
# Update the feature hash ranges (log-2 units)
lgbm_param <- lgbm_wflow |>
extract_parameter_set_dials() |>
update(
`agent hash` = num_hash(c(3, 8)),
`company hash` = num_hash(c(3, 8))
)
# tune the model
lgbm_res <- lgbm_wflow |>
tune_grid(
resamples = hotel_rs,
grid = 25,
# The options below are not required by default
param_info = lgbm_param,
control = ctrl,
metrics = reg_metrics
)
# A tibble: 5 × 11
min_n learn_rate stop_iter `agent hash` `company hash` .metric .estimator
<int> <dbl> <int> <int> <int> <chr> <chr>
1 9 0.0712 12 61 28 mae standard
2 12 0.0180 6 13 9 mae standard
3 30 0.0409 13 37 44 mae standard
4 24 0.00495 4 92 28 mae standard
5 33 0.00200 14 23 11 mae standard
# ℹ 4 more variables: mean <dbl>, n <int>, std_err <dbl>, .config <chr>Our boosting model
We used feature hashing to generate a smaller set of indicator columns to deal with the large number of levels for the agent and country predictors.
Tree-based models (and a few others) don’t require indicators for categorical predictors. They can split on these variables as-is.
We’ll keep all categorical predictors as factors and focus on optimizing additional boosting parameters.
Our Boosting Model
lgbm_spec <- boost_tree(
trees = 1000, learn_rate = tune(), min_n = tune(),
tree_depth = tune(), loss_reduction = tune(),
stop_iter = tune()
) |>
set_mode("regression") |>
set_engine("lightgbm")
lgbm_wflow <- workflow(avg_price_per_room ~ ., lgbm_spec)
lgbm_param <- lgbm_wflow |>
extract_parameter_set_dials() |>
update(learn_rate = learn_rate(c(-5, -1)))Iterative search
Instead of pre-defining a grid of candidate points, we can model our current results to predict what the next candidate point should be.
Suppose that we are only tuning the learning rate in our boosted tree.
Iterative Search
A linear model probably isn’t the best choice though (more in a minute).
To illustrate the process, we resampled a large grid of learning rate values for our data to show what the relationship is between MAE and learning rate.
Now suppose that we used a grid of three points in the parameter range for learning rate…
A large grid

A three point grid

Bayesian optimization
- A sequential method that uses a model to predict new candidate parameters for assessment
- When scoring potential parameter value, the mean and variance of performance are predicted
- The strategy used to define how these two statistical quantities are used is defined by an acquisition function
Acquisition function
Acquisition functions take the predicted mean and variance and use them to balance:
- exploration: new candidates should explore new areas.
- exploitation: new candidates must stay near existing values.
Exploration focuses on the variance, exploitation is about the mean.
Acquisition functions
We’ll use an acquisition function to select a new candidate.
The most popular method appears to be expected improvement (EI) above the current best results.
- Zero at existing data points.
- The expected improvement is integrated over all possible improvement (“expected” in the probability sense).
We would probably pick the point with the largest EI as the next point.
(There are other functions beyond EI.)
Iteration
Once we pick the candidate point, we measure performance for it (e.g. resampling).
Another GP is fit, EI is recomputed, and so on.
We stop when we have completed the allowed number of iterations or if we don’t see any improvement after a pre-set number of attempts.
GP evolution
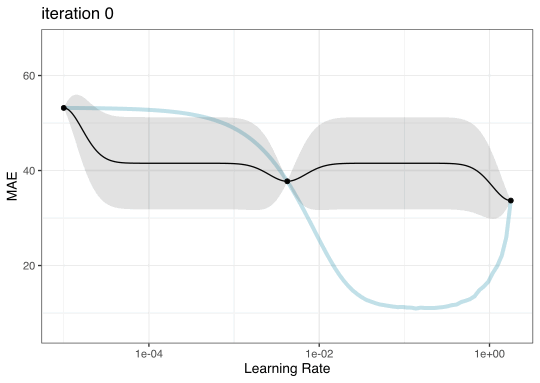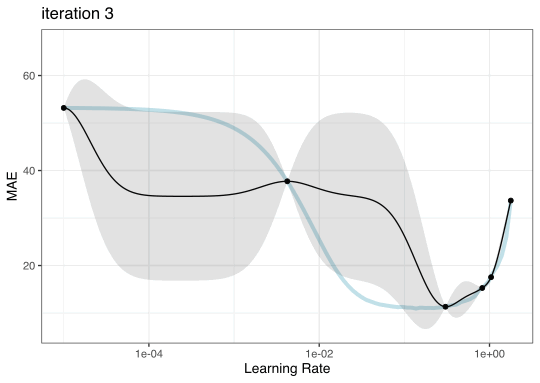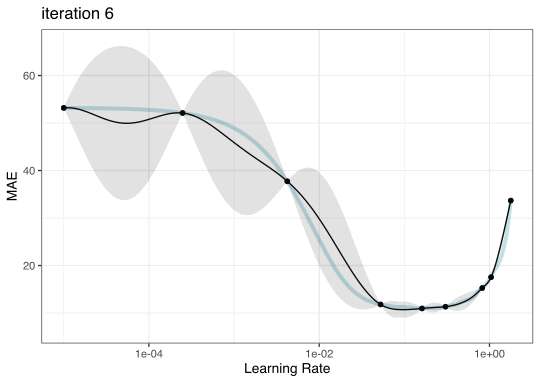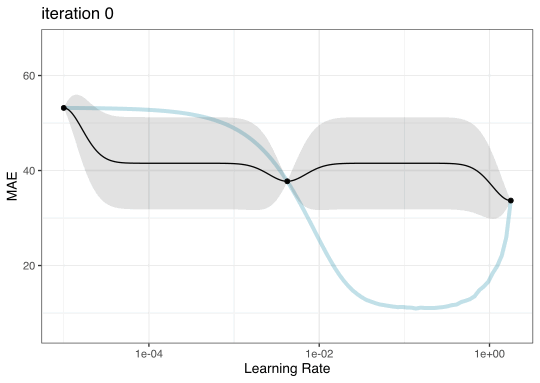
BO in {tidymodels}
We’ll use a function called tune_bayes() that has very similar syntax to tune_grid().
It has an additional initial argument for the initial set of performance estimates and parameter combinations for the GP model.
Initial grid points
initial can be the results of another tune_*() function or an integer (in which case tune_grid() is used under to hood to make such an initial set of results).
- We’ll run the optimization more than once, so let’s make an initial grid of results to serve as the substrate for the BO.
An initial grid
reg_metrics <- metric_set(mae, rsq)
set.seed(12)
init_res <- lgbm_wflow |>
tune_grid(
resamples = hotel_rs,
grid = nrow(lgbm_param) + 2,
param_info = lgbm_param,
metrics = reg_metrics
)
show_best(init_res, metric = "mae") |> select(-.metric, -.estimator)# A tibble: 5 × 9
min_n tree_depth learn_rate loss_reduction stop_iter mean n std_err
<int> <int> <dbl> <dbl> <int> <dbl> <int> <dbl>
1 9 4 0.0415 5.21e- 9 13 9.96 10 0.176
2 16 12 0.0136 1.91e- 3 9 10.1 10 0.156
3 25 8 0.00256 9.58e-10 7 14.2 10 0.174
4 22 9 0.00154 5.77e- 6 5 19.2 10 0.170
5 32 3 0.000144 3.02e+ 1 18 47.9 10 0.302
# ℹ 1 more variable: .config <chr>BO using {tidymodels}
ctrl_bo <- control_bayes(verbose_iter = TRUE) # <- for demonstration
set.seed(15)
lgbm_bayes_res <- lgbm_wflow |>
tune_bayes(
resamples = hotel_rs,
initial = init_res, # <- initial results
iter = 20,
param_info = lgbm_param,
control = ctrl_bo,
metrics = reg_metrics
)Optimizing mae using the expected improvement── Iteration 1 ─────────────────────────────────────────────────────────────────i Current best: mae=9.964 (@iter 0)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=32, tree_depth=12, learn_rate=0.0178, loss_reduction=1.03e-10,
stop_iter=12i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.02 (+/-0.155)── Iteration 2 ─────────────────────────────────────────────────────────────────i Current best: mae=9.964 (@iter 0)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=15, tree_depth=14, learn_rate=0.0977, loss_reduction=0.00535,
stop_iter=4i Estimating performance✓ Estimating performance♥ Newest results: mae=9.707 (+/-0.167)── Iteration 3 ─────────────────────────────────────────────────────────────────i Current best: mae=9.707 (@iter 2)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=15, tree_depth=2, learn_rate=0.0986, loss_reduction=2.65,
stop_iter=20i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.86 (+/-0.177)── Iteration 4 ─────────────────────────────────────────────────────────────────i Current best: mae=9.707 (@iter 2)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=12, tree_depth=12, learn_rate=0.0308, loss_reduction=2.16e-06,
stop_iter=3i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.801 (+/-0.142)── Iteration 5 ─────────────────────────────────────────────────────────────────i Current best: mae=9.707 (@iter 2)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=37, tree_depth=15, learn_rate=0.0501, loss_reduction=1.89,
stop_iter=9i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.807 (+/-0.155)── Iteration 6 ─────────────────────────────────────────────────────────────────i Current best: mae=9.707 (@iter 2)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=9, tree_depth=14, learn_rate=0.0624, loss_reduction=0.000108,
stop_iter=3i Estimating performance✓ Estimating performance♥ Newest results: mae=9.567 (+/-0.145)── Iteration 7 ─────────────────────────────────────────────────────────────────i Current best: mae=9.567 (@iter 6)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=11, tree_depth=2, learn_rate=0.0441, loss_reduction=0.0203,
stop_iter=3i Estimating performance✓ Estimating performanceⓧ Newest results: mae=11.46 (+/-0.203)── Iteration 8 ─────────────────────────────────────────────────────────────────i Current best: mae=9.567 (@iter 6)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=13, tree_depth=15, learn_rate=0.0385, loss_reduction=2.86e-09,
stop_iter=18i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.64 (+/-0.139)── Iteration 9 ─────────────────────────────────────────────────────────────────i Current best: mae=9.567 (@iter 6)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=8, tree_depth=8, learn_rate=0.064, loss_reduction=1.22e-06,
stop_iter=20i Estimating performance✓ Estimating performance♥ Newest results: mae=9.55 (+/-0.151)── Iteration 10 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=39, tree_depth=7, learn_rate=0.0888, loss_reduction=1.36e-10,
stop_iter=9i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.855 (+/-0.157)── Iteration 11 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=17, tree_depth=13, learn_rate=0.0683, loss_reduction=3.25e-10,
stop_iter=11i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.65 (+/-0.161)── Iteration 12 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=6, tree_depth=6, learn_rate=0.0102, loss_reduction=9.86e-10,
stop_iter=19i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.4 (+/-0.189)── Iteration 13 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=4, tree_depth=11, learn_rate=0.0414, loss_reduction=0.00021,
stop_iter=15i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.567 (+/-0.16)── Iteration 14 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=19, tree_depth=14, learn_rate=0.00693, loss_reduction=0.0704,
stop_iter=19i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.63 (+/-0.185)── Iteration 15 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=2, tree_depth=11, learn_rate=0.0701, loss_reduction=7.25e-06,
stop_iter=19i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.581 (+/-0.15)── Iteration 16 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=2, tree_depth=6, learn_rate=0.0631, loss_reduction=26.7, stop_iter=17i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.16 (+/-0.171)── Iteration 17 ────────────────────────────────────────────────────────────────i Current best: mae=9.55 (@iter 9)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=4, tree_depth=13, learn_rate=0.0524, loss_reduction=9.64e-09,
stop_iter=13i Estimating performance✓ Estimating performance♥ Newest results: mae=9.547 (+/-0.173)── Iteration 18 ────────────────────────────────────────────────────────────────i Current best: mae=9.547 (@iter 17)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=16, tree_depth=12, learn_rate=0.04, loss_reduction=6.77e-07,
stop_iter=17i Estimating performance✓ Estimating performanceⓧ Newest results: mae=9.611 (+/-0.149)── Iteration 19 ────────────────────────────────────────────────────────────────i Current best: mae=9.547 (@iter 17)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=17, tree_depth=15, learn_rate=0.0107, loss_reduction=1.33e-09,
stop_iter=3i Estimating performance✓ Estimating performanceⓧ Newest results: mae=10.24 (+/-0.155)── Iteration 20 ────────────────────────────────────────────────────────────────i Current best: mae=9.547 (@iter 17)i Gaussian process model✓ Gaussian process modeli Generating 5000 candidatesi Predicted candidatesi min_n=4, tree_depth=10, learn_rate=0.0592, loss_reduction=1.99e-10,
stop_iter=11i Estimating performance✓ Estimating performance♥ Newest results: mae=9.493 (+/-0.161)Best results
# A tibble: 5 × 10
min_n tree_depth learn_rate loss_reduction stop_iter mean n std_err
<int> <int> <dbl> <dbl> <int> <dbl> <int> <dbl>
1 4 10 0.0592 1.99e-10 11 9.49 10 0.161
2 4 13 0.0524 9.64e- 9 13 9.55 10 0.173
3 8 8 0.0640 1.22e- 6 20 9.55 10 0.151
4 4 11 0.0414 2.10e- 4 15 9.57 10 0.160
5 9 14 0.0624 1.08e- 4 3 9.57 10 0.145
# ℹ 2 more variables: .config <chr>, .iter <int>Plotting BO results

Plotting BO results

Plotting BO results

ENHANCE

Notes
Stopping
tune_bayes()will return the current results.Parallel processing can still be used to more efficiently measure each candidate point.
There are a lot of other iterative methods that you can use.
The {finetune} package also has functions for simulated annealing search.
Finalizing the model
Finalizing the model
Let’s say that we’ve tried a lot of different models and we like our LightGBM model the most.
What do we do now?
- Finalize the workflow by choosing the values for the tuning parameters.
- Fit the model on the entire training set.
- Verify performance using the test set.
- Document and publish the model (later this semester)
Locking down the tuning parameters
We can take the results of the Bayesian optimization and accept the best results:
best_param <- select_best(lgbm_bayes_res, metric = "mae")
final_wflow <- lgbm_wflow |>
finalize_workflow(best_param)
final_wflow══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Formula
Model: boost_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
avg_price_per_room ~ .
── Model ───────────────────────────────────────────────────────────────────────
Boosted Tree Model Specification (regression)
Main Arguments:
trees = 1000
min_n = 4
tree_depth = 10
learn_rate = 0.0592446327108508
loss_reduction = 1.9888406080826e-10
stop_iter = 11
Computational engine: lightgbm The final fit
We can use individual functions:
final_fit <- final_wflow |> fit(data = hotel_train)
# then predict() or augment()
# then compute metricsRemember that there is also a convenience function to do all of this:
# Resampling results
# Manual resampling
# A tibble: 1 × 6
splits id .metrics .notes .predictions .workflow
<list> <chr> <list> <list> <list> <list>
1 <split [3749/1251]> train/test split <tibble> <tibble> <tibble> <workflow>Test set results
Test set performance:

Recall that resampling predicted the MAE to be 9.49.
Wrap-up
Recap
- Tuning parameters significantly impact the performance of models
- Tuning parameters can be defined in both the feature engineering and model specification stages
- Grid search is a simple but effective method for tuning parameters
- Iterative search methods can be used to optimize parameters
- Bayesian optimization is a sequential method that uses a model to predict new candidate parameters for assessment
- The {tidymodels} framework provides functions for grid search and Bayesian optimization
Acknowledgments
- Materials derived in part from Machine learning with {tidymodels} and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.
Apple Harvest Festival


