# A tibble: 7,107 × 20
forested year elevation eastness northness roughness tree_no_tree dew_temp precip_annual
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <dbl> <dbl>
1 Yes 2005 881 90 43 63 Tree 0.04 466
2 Yes 2005 113 -25 96 30 Tree 6.4 1710
3 No 2005 164 -84 53 13 Tree 6.06 1297
4 Yes 2005 299 93 34 6 No tree 4.43 2545
5 Yes 2005 806 47 -88 35 Tree 1.06 609
6 Yes 2005 736 -27 -96 53 Tree 1.35 539
7 Yes 2005 636 -48 87 3 No tree 1.42 702
8 Yes 2005 224 -65 -75 9 Tree 6.39 1195
9 Yes 2005 52 -62 78 42 Tree 6.5 1312
10 Yes 2005 2240 -67 -74 99 No tree -5.63 1036
# ℹ 7,097 more rows
# ℹ 11 more variables: temp_annual_mean <dbl>, temp_annual_min <dbl>, temp_annual_max <dbl>,
# temp_january_min <dbl>, vapor_min <dbl>, vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>,
# lat <dbl>, land_type <fct>, county <fct>Evaluate models using appropriate metrics
Lecture 8
Cornell University
INFO 4940/5940 - Fall 2025
September 18, 2025
Announcements
Announcements
- Homework 03
- Project 1 coming soon
Learning objectives
- Define metrics for evaluating the performance of classification and regression models
- Identify trade-offs between different metrics
- Interpret different metrics
- Select appropriate metrics for a given modeling task
Data on forests in Washington
Data on forests in Washington
- The U.S. Forest Service maintains ML models to predict whether a plot of land is “forested.”
- This classification is important for all sorts of research, legislation, and land management purposes.
- Plots are typically remeasured every 10 years and this dataset contains the most recent measurement per plot.
Data on forests in Washington
N = 7,107plots of land, one from each of 7,107 6000-acre hexagons in WA.- A nominal outcome,
forested, with levels"Yes"and"No", measured “on-the-ground.” - 18 remotely-sensed and easily-accessible predictors:
- numeric variables based on weather and topography.
- nominal variables based on classifications from other governmental orgs.
Data on forests in Washington
Partition and fit a very basic model
Show the code
library(tidymodels)
library(forested)
set.seed(123)
forested_split <- initial_split(forested, prop = 0.8)
forested_train <- training(forested_split)
forested_test <- testing(forested_split)
tree_spec <- decision_tree(cost_complexity = 0.0001, mode = "classification")
forested_wflow <- workflow(forested ~ ., tree_spec)
forested_fit <- fit(forested_wflow, forested_train)Looking at predictions
Looking at predictions
# A tibble: 1,422 × 23
.pred_class .pred_Yes .pred_No forested year elevation eastness northness roughness tree_no_tree
<fct> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 Yes 0.977 0.0226 No 2005 164 -84 53 13 Tree
2 Yes 0.833 0.167 Yes 2005 299 93 34 6 No tree
3 No 0.154 0.846 Yes 2005 636 -48 87 3 No tree
4 No 0.25 0.75 Yes 2005 224 -65 -75 9 Tree
5 No 0.263 0.737 Yes 2005 2240 -67 -74 99 No tree
6 Yes 0.977 0.0226 Yes 2004 1044 96 -26 51 Tree
7 Yes 0.977 0.0226 Yes 2003 1031 -49 86 190 Tree
8 Yes 0.977 0.0226 Yes 2005 1330 99 7 212 Tree
9 Yes 0.977 0.0226 Yes 2005 1423 46 88 180 Tree
10 Yes 0.977 0.0226 Yes 2014 546 -92 -38 28 Tree
# ℹ 1,412 more rows
# ℹ 13 more variables: dew_temp <dbl>, precip_annual <dbl>, temp_annual_mean <dbl>,
# temp_annual_min <dbl>, temp_annual_max <dbl>, temp_january_min <dbl>, vapor_min <dbl>,
# vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>, land_type <fct>, county <fct>Confusion matrix
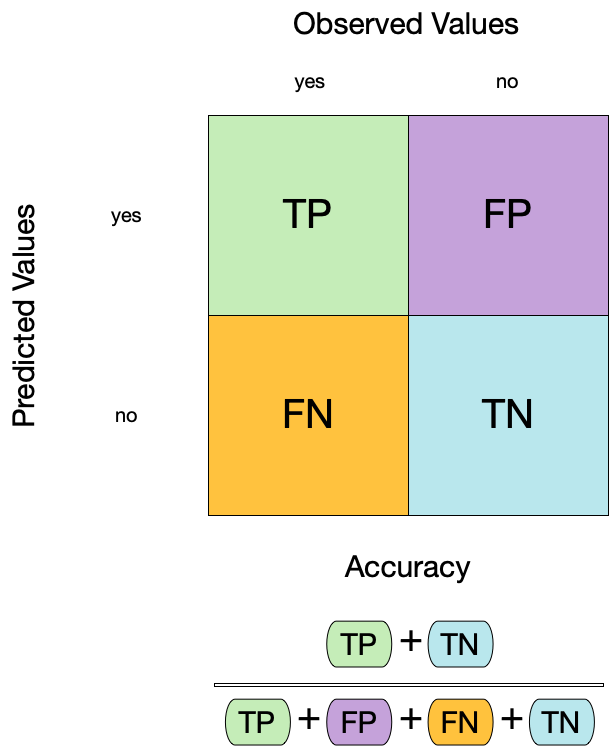
Confusion matrix
Confusion matrix
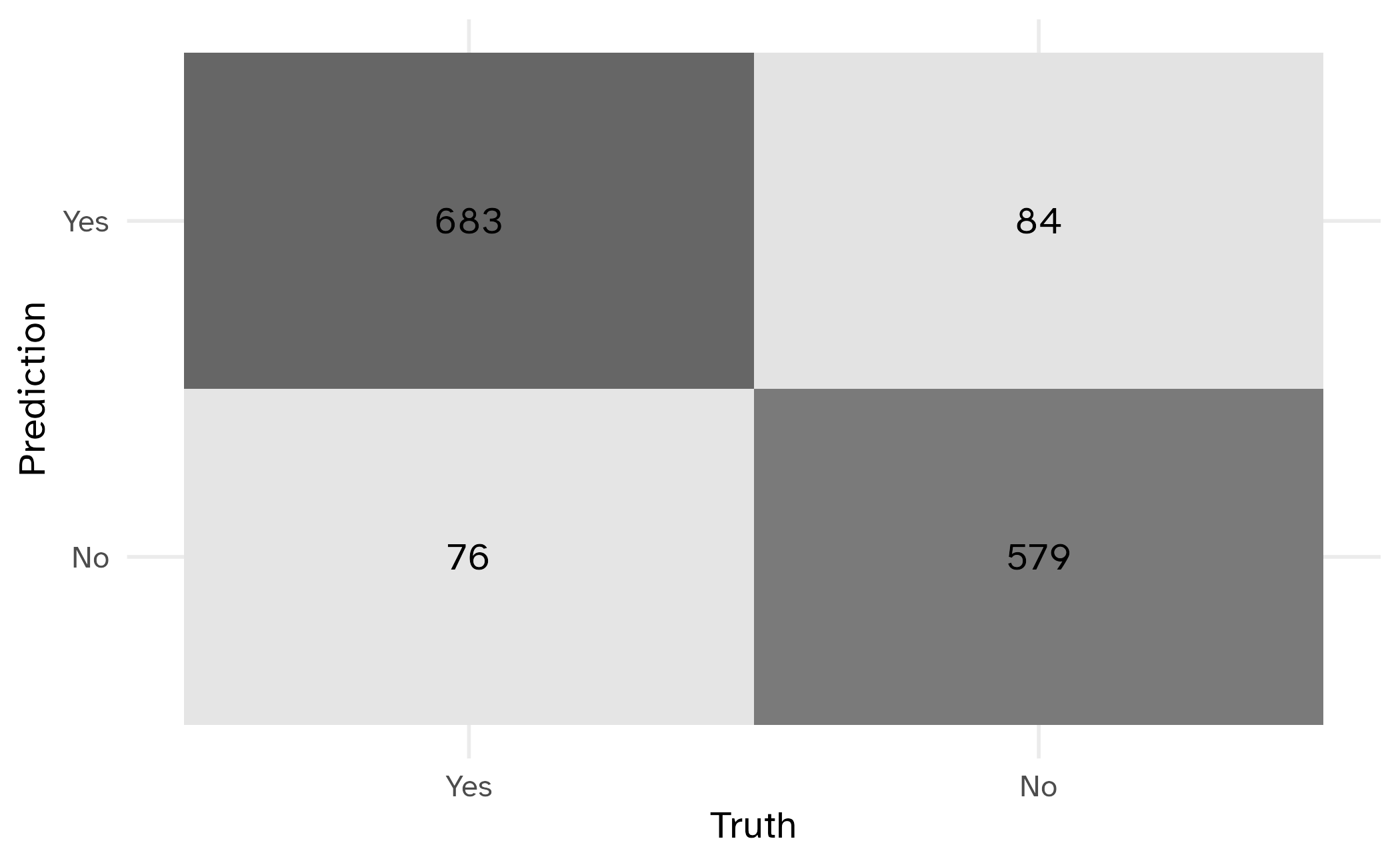
Metrics for model performance
Metrics for model performance
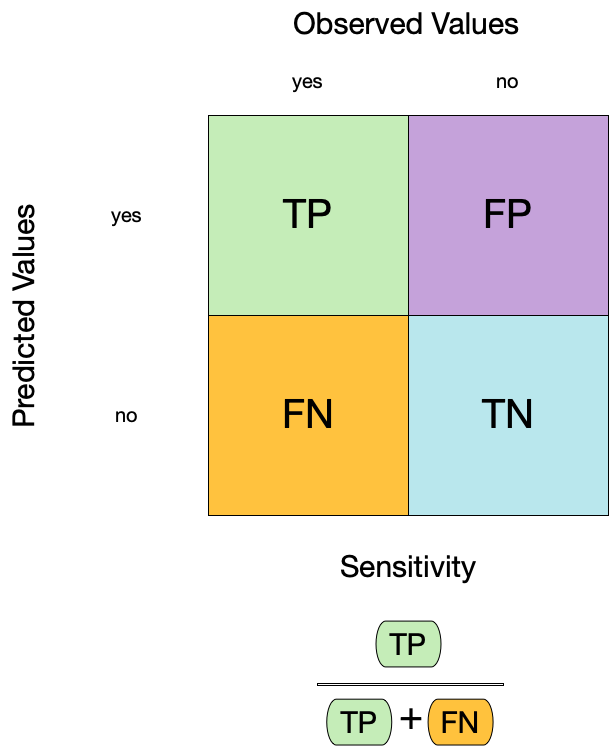
Sensitivity (TPR)
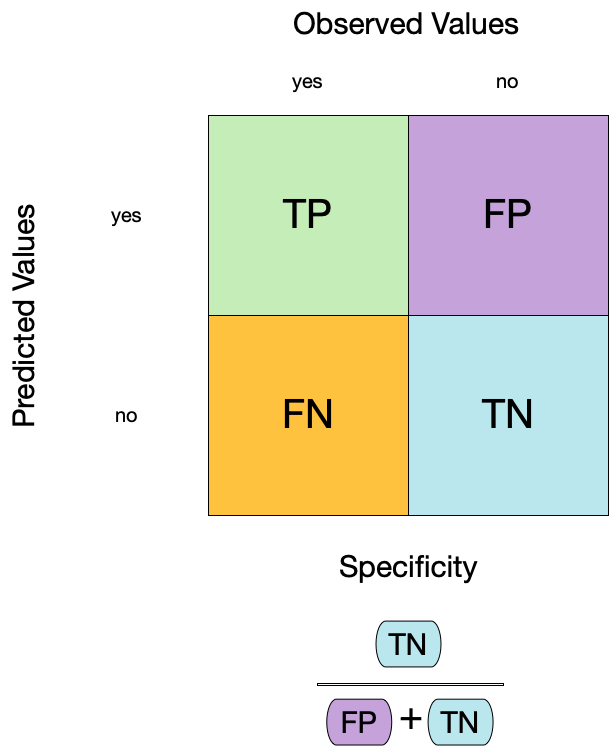
Specificity (TNR)
Metrics for model performance
Show the code
# A tibble: 3 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.887
2 specificity binary 0.873
3 sensitivity binary 0.900Group differences in metrics
📝 Group differences in metrics
Instructions
Do any metrics differ substantially between groups? What does this mean?
# A tibble: 6 × 4
tree_no_tree .metric .estimator .estimate
<fct> <chr> <chr> <dbl>
1 Tree accuracy binary 0.899
2 No tree accuracy binary 0.876
3 Tree specificity binary 0.372
4 No tree specificity binary 0.940
5 Tree sensitivity binary 0.964
6 No tree sensitivity binary 0.583
03:00
Combined metrics
\(J\) index (combines sensitivity and specificity)
\[ J = \text{Sensitivity} + \text{Specificity} - 1 \]
\(F\)-score (combines precision and recall)
\[ F_{1}=2\left(\frac{\text{Precision} \times \text{Recall}}{\text{Precision} + \text{Recall}}\right) \]
Two class data
These metrics assume that we know the threshold for converting “soft” probability predictions into “hard” class predictions.
Is a 50% threshold good?
What happens if we say that we need to be 80% sure to declare an event?
- sensitivity ⬇️, specificity ⬆️
What happens for a 20% threshold?
- sensitivity ⬆️, specificity ⬇️
Varying the threshold
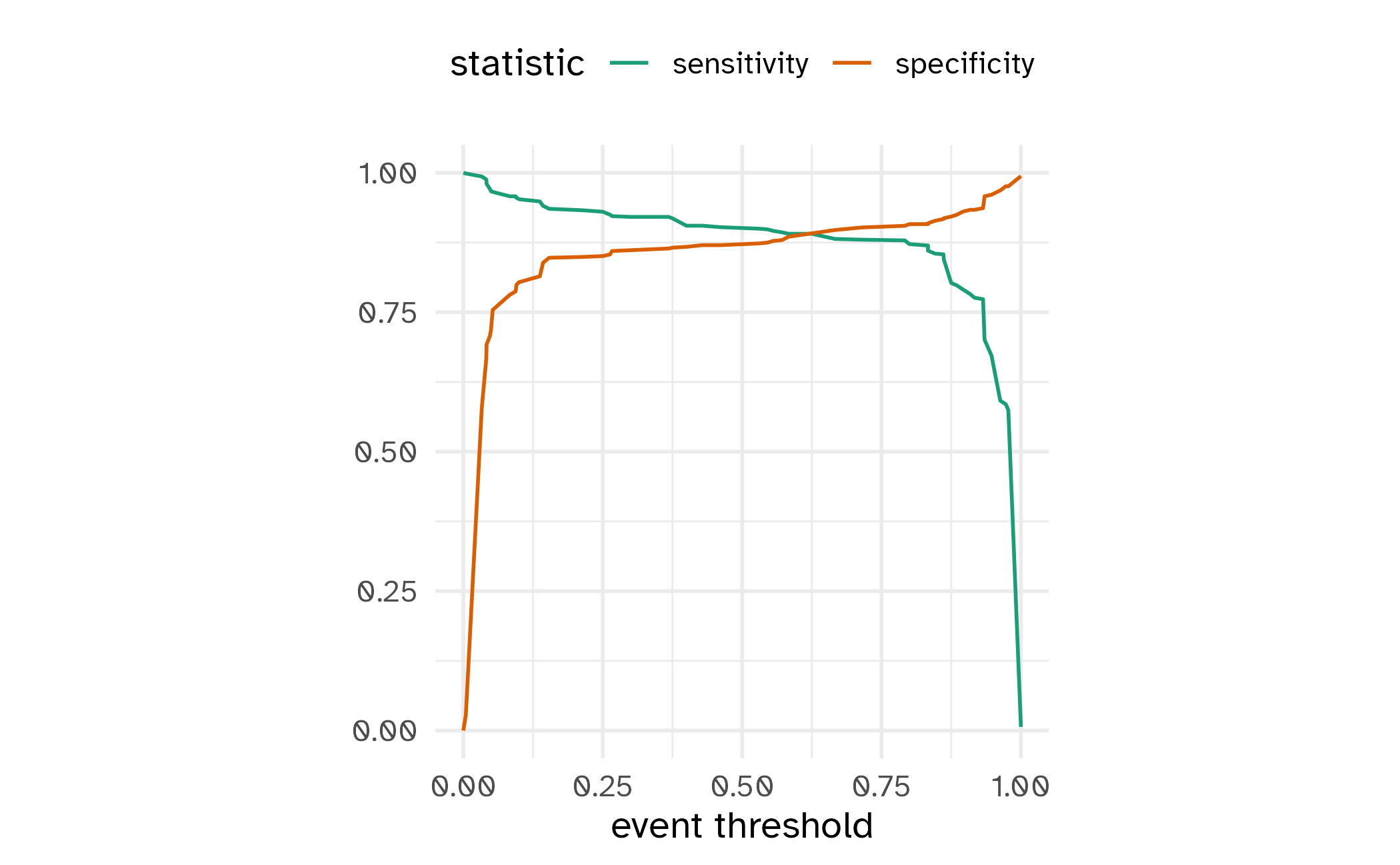
ROC curves
For an ROC (receiver operator characteristic) curve, we plot
- the false positive rate (1 - specificity) on the x-axis
- the true positive rate (sensitivity) on the y-axis
with sensitivity and specificity calculated at all possible thresholds.
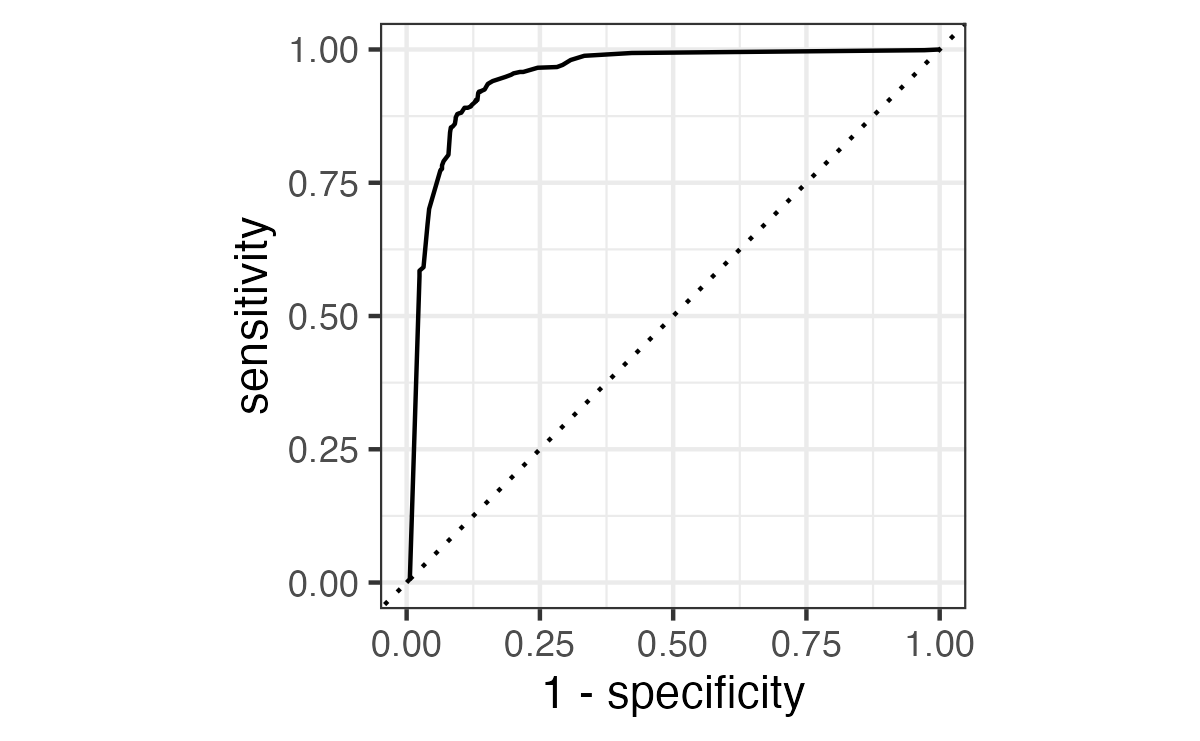
ROC curves
We can use the area under the ROC curve as a classification metric:
- ROC AUC = 1 💯
- ROC AUC = 1/2 😢
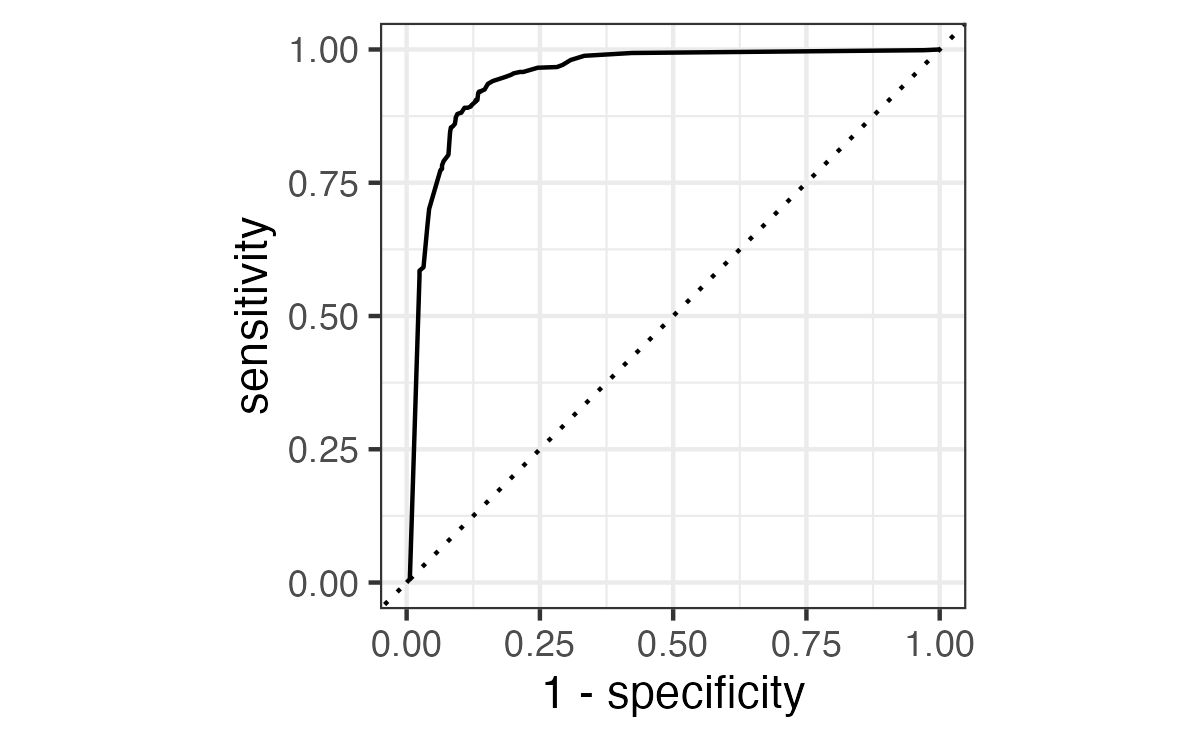
ROC curves
Show the code
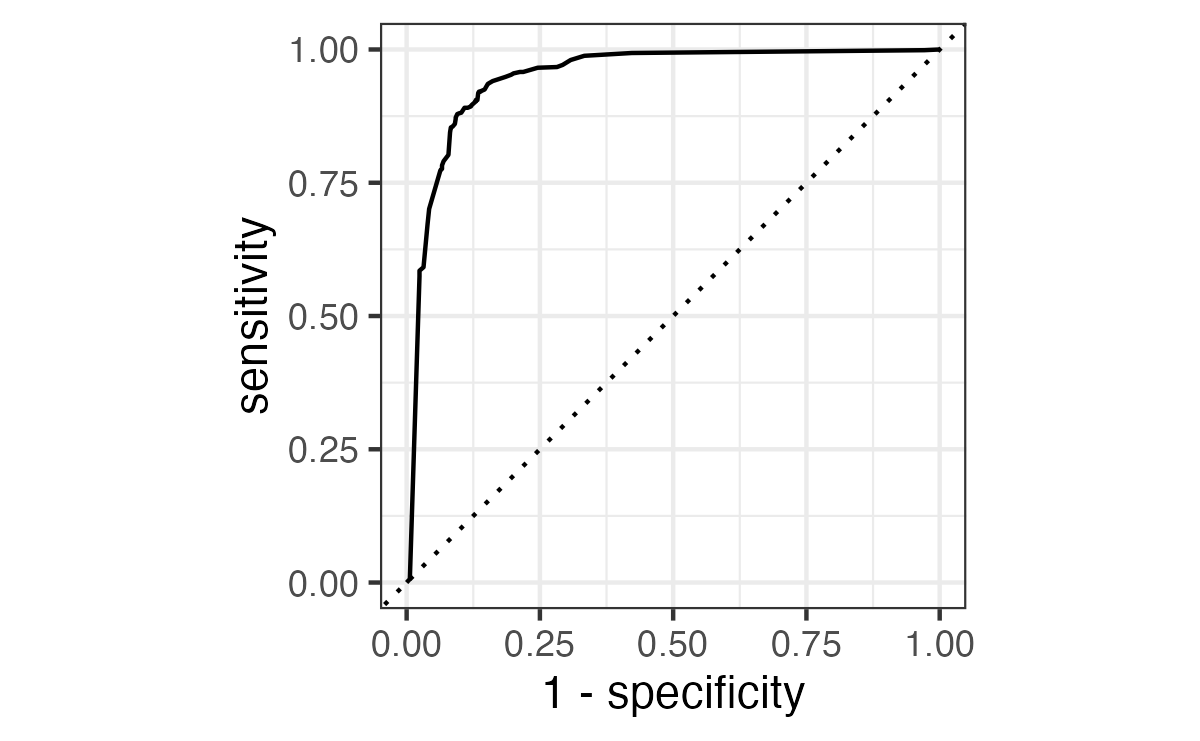
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.948Brier score
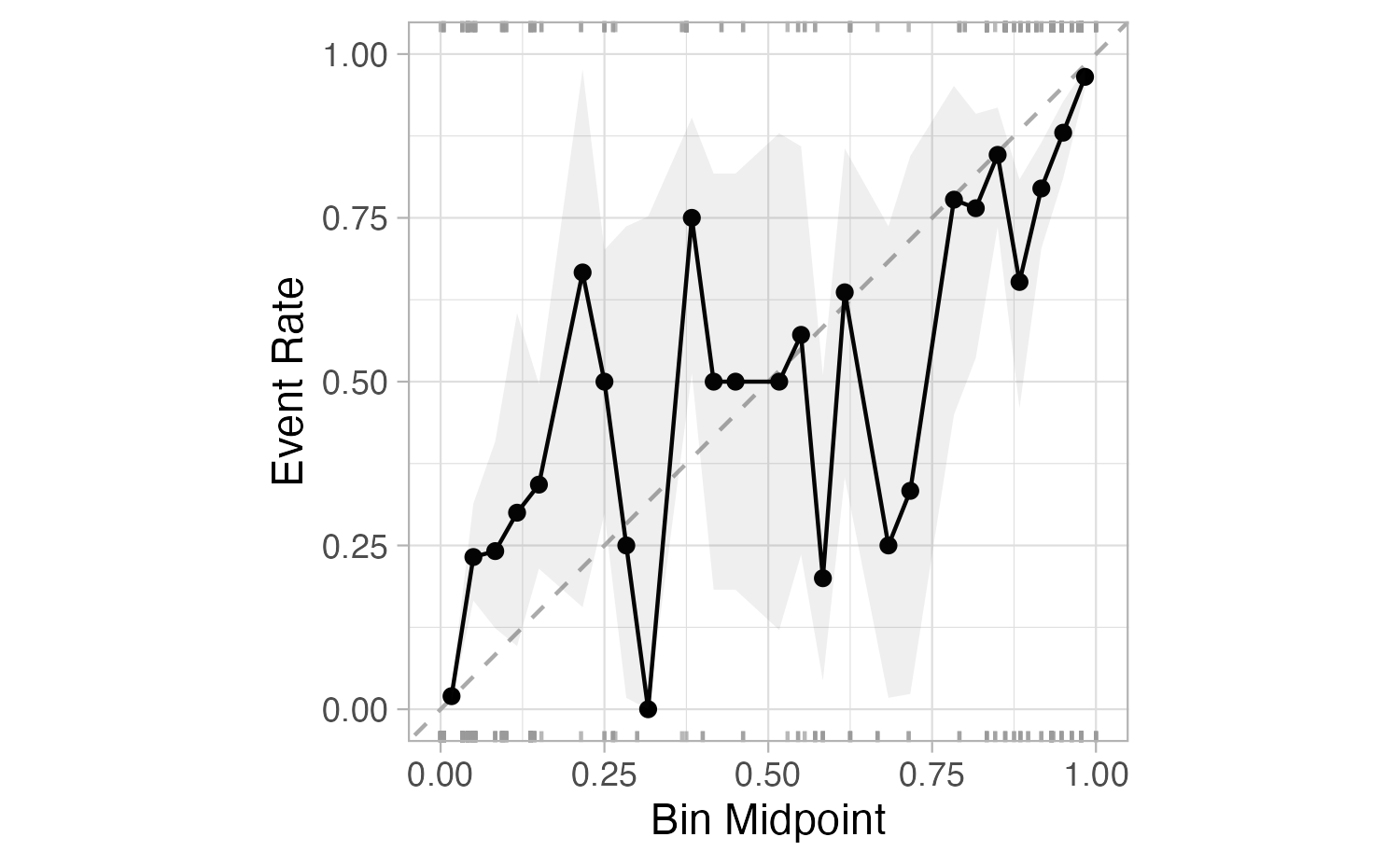
What if we don’t turn predicted probabilities into class predictions?
The Brier score is analogous to the mean squared error in regression models:
\[ \text{Brier}_{\text{class}} = \frac{1}{N}\sum_{i=1}^N\sum_{k=1}^C (y_{ik} - \hat{p}_{ik})^2 \]
Brier score
Show the code
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 brier_class binary 0.0877Smaller values are better, for binary classification the “bad model threshold” is about 0.25.
Separation vs calibration
The ROC captures separation
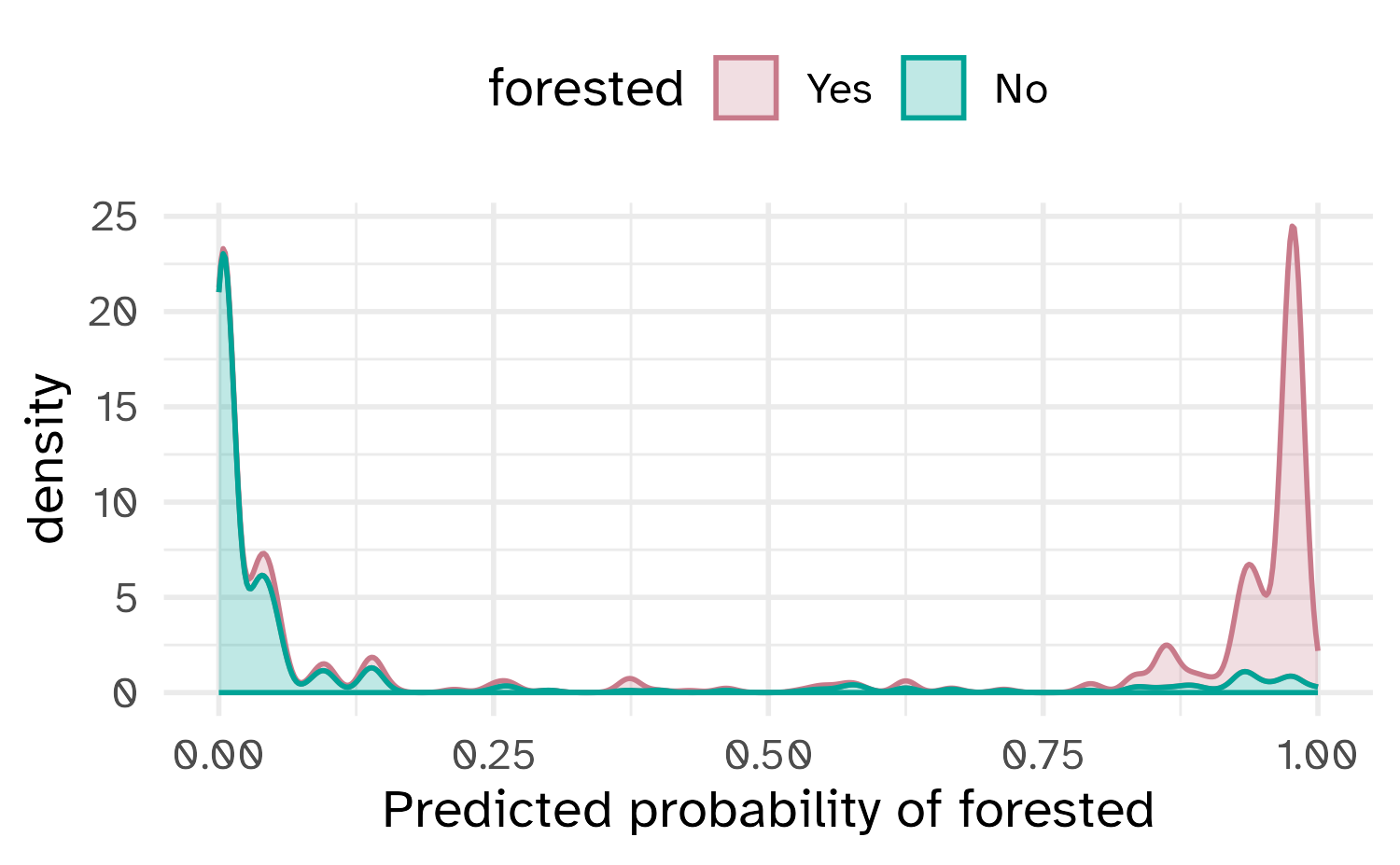
The Brier score captures calibration
Common metrics for regression outcomes
Absolute error metrics
Expressed in the same units as the outcome of interest
Root mean squared error (RMSE)
\[ \text{RMSE} = \sqrt{\frac{1}{n}\sum_{i=1}^{n}(y_i - \hat{y}_i)^2} \]
Mean absolute error (MAE)
\[ \text{MAE} = \frac{1}{n}\sum_{i=1}^{n}|y_i - \hat{y}_i| \]
RMSE is more sensitive to large errors than MAE
Percentage/relative error metrics
Expressed in relative terms, e.g. as a percentage of the actual value
Mean percentage error
\[ \text{MPE} = 100 \times \frac{1}{n}\sum_{i=1}^{n}\frac{y_i - \hat{y}_i}{y_i} \]
Mean absolute percent error
\[ \text{MAPE} = 100 \times \frac{1}{n}\sum_{i=1}^{n}\left|\frac{y_i - \hat{y}_i}{y_i}\right| \]
Equivalent to absolute error metrics, expressed in relative terms
Correlation metrics
Measure of correlation/consistency between predicted and actual values
R-squared
\[ R^2 = 1 - \frac{\sum_{i=1}^{n}(y_i - \hat{y}_i)^2}{\sum_{i=1}^{n}(y_i - \bar{y})^2} \]
Ranges from 0 to 1, higher values are better
Concordance correlation coefficient
\[ \rho_c = \frac{2\rho\sigma_y\sigma_\hat{y}}{\sigma_y^2 + \sigma_\hat{y}^2 + (\mu_y - \mu_\hat{y})^2} \]
Evaluates both the accuracy and consistency of predictions
Selecting appropriate metrics
Selecting appropriate metrics
Understand the type of problem
- Classification (binary or multiclass)
- Regression
- Survival analysis
- Clustering
Define objectives and prioritize errors
- What stakeholders care about
- Consequences of different types of errors
Consider data characteristics
- Class imbalance
- Outliers
- Data distribution
Use multiple metrics (where possible)
- Use a combined metric (e.g. \(F\)-score, \(J\)-index)
- Desirability functions with {desirability2}
📝 Selecting appropriate metrics
Instructions
You’re evaluating a diagnostic model for COVID-19 detection based on symptoms (fever, cough, loss of taste/smell, fatigue, etc.) and rapid saliva testing.
True Positive: Model correctly identifies COVID → Patient isolates, gets treatment, contact tracing begins
False Positive: Model incorrectly flags healthy person as COVID → Unnecessary isolation, anxiety, lost work
True Negative: Model correctly identifies non-COVID illness → Patient gets appropriate care, no isolation
False Negative: Model misses COVID case → Patient spreads virus, no isolation, delayed treatment
| Stakeholder | Most Important Metric | Least Acceptable Error Type | Why? |
|---|---|---|---|
| Individual Patient | |||
| Hospital administrator | |||
| Public Health Official | |||
| Workplace/School |
10:00
Wrap-up
Recap
- Metrics are used to evaluate model performance
- Different metrics capture different aspects of performance
- Select metrics that align with your goals and stakeholders
Acknowledgments
- Materials derived in part from Machine learning with {tidymodels} and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.