These are suggested answers. This document should be used as reference only, it’s not designed to be an exhaustive key.
Data on forests in Washington
── Attaching packages ────────────────────────────────────── tidymodels 1.2.0 ──
✔ broom 1.0.6 ✔ recipes 1.0.10
✔ dials 1.3.0 ✔ rsample 1.2.1
✔ dplyr 1.1.4 ✔ tibble 3.2.1
✔ ggplot2 3.5.1 ✔ tidyr 1.3.1
✔ infer 1.0.7 ✔ tune 1.2.1
✔ modeldata 1.4.0 ✔ workflows 1.1.4
✔ parsnip 1.2.1 ✔ workflowsets 1.1.0
✔ purrr 1.0.2 ✔ yardstick 1.3.1
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ purrr::%||%() masks base::%||%()
✖ purrr::discard() masks scales::discard()
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
✖ recipes::step() masks stats::step()
• Dig deeper into tidy modeling with R at https://www.tmwr.org
# A tibble: 7,107 × 19
forested year elevation eastness northness roughness tree_no_tree dew_temp
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <dbl>
1 Yes 2005 881 90 43 63 Tree 0.04
2 Yes 2005 113 -25 96 30 Tree 6.4
3 No 2005 164 -84 53 13 Tree 6.06
4 Yes 2005 299 93 34 6 No tree 4.43
5 Yes 2005 806 47 -88 35 Tree 1.06
6 Yes 2005 736 -27 -96 53 Tree 1.35
7 Yes 2005 636 -48 87 3 No tree 1.42
8 Yes 2005 224 -65 -75 9 Tree 6.39
9 Yes 2005 52 -62 78 42 Tree 6.5
10 Yes 2005 2240 -67 -74 99 No tree -5.63
# ℹ 7,097 more rows
# ℹ 11 more variables: precip_annual <dbl>, temp_annual_mean <dbl>,
# temp_annual_min <dbl>, temp_annual_max <dbl>, temp_january_min <dbl>,
# vapor_min <dbl>, vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>,
# land_type <fct>
Your turn
When is a good time to split your data?
Add response here After all your data sources are combined together before you attempt any modeling procedures.
Data splitting and spending
set.seed ( 123 ) forested_split <- initial_split ( forested ) forested_split
<Training/Testing/Total>
<5330/1777/7107>
Extract the training and testing sets
forested_train <- training ( forested_split ) forested_test <- testing ( forested_split )
Your turn
Split your data so 20% is held out for the test set.
Try out different values in set.seed()
Hint: Which argument in initial_split() handles the proportion split into training vs testing?
set.seed ( 123 ) forested_split <- initial_split ( forested , prop = 0.8 ) forested_split
<Training/Testing/Total>
<5685/1422/7107>
set.seed ( 456 ) forested_split <- initial_split ( forested , prop = 0.8 ) forested_split
<Training/Testing/Total>
<5685/1422/7107>
set.seed ( 789 ) forested_split <- initial_split ( forested , prop = 0.8 ) forested_split
<Training/Testing/Total>
<5685/1422/7107>
Exploratory data analysis
Explore the forested_train data on your own!
What’s the distribution of the outcome, forested?
What’s the distribution of numeric variables like precip_annual?
How does the distribution of forested differ across the categorical variables?
# bar chart of forested forested_train |> ggplot ( aes ( x = forested ) ) +
geom_bar ( ) # forested vs. tree/no tree forested_train |> ggplot ( aes ( x = forested , fill = tree_no_tree ) ) +
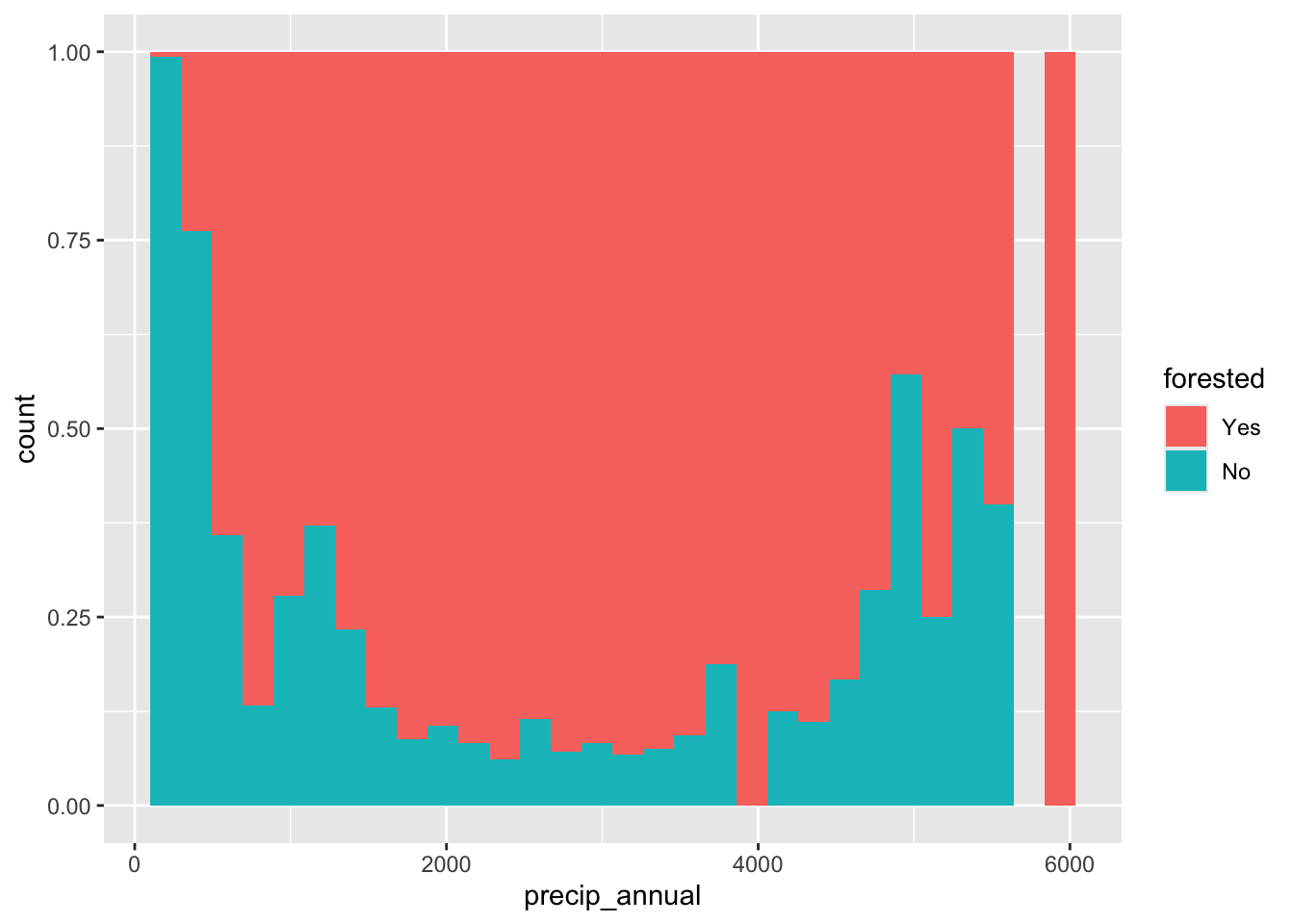
geom_bar ( ) # forested vs. precipitation forested_train |> ggplot ( aes ( x = precip_annual , fill = forested , color = forested ) ) +
geom_freqpoly ( bins = 30 ) forested_train |> ggplot ( aes ( x = precip_annual , fill = forested , group = forested ) ) +
geom_histogram ( position = "fill" )
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).
# forested by location forested_train |> ggplot ( aes ( x = lon , y = lat , col = forested ) ) +
geom_point ( )
Fitting models
To specify a model
Linear Regression Model Specification (regression)
Computational engine: lm
# Engine linear_reg ( ) |> set_engine ( "glmnet" )
Linear Regression Model Specification (regression)
Computational engine: glmnet
# Mode - Some models have a default mode, others don't decision_tree ( ) |> set_mode ( "regression" )
Decision Tree Model Specification (regression)
Computational engine: rpart
Your turn
Edit the chunk below to use a logistic regression model.
Extension/Challenge: Edit this code to use a different model. For example, try using a conditional inference tree as implemented in the partykit package by changing the engine - or try an entirely different model type!
All available models are listed at https://www.tidymodels.org/find/parsnip/
tree_spec <- decision_tree ( ) |> set_mode ( "classification" )
tree_spec
Decision Tree Model Specification (classification)
Computational engine: rpart
log_spec <- logistic_reg ( ) |> set_mode ( "classification" )
log_spec
Logistic Regression Model Specification (classification)
Computational engine: glm
A model workflow
tree_spec <- decision_tree ( ) |> set_mode ( "classification" )
Fit parsnip specification:
tree_spec |> fit ( forested ~ . , data = forested_train )
parsnip model object
n= 5330
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5330 2398 Yes (0.55009381 0.44990619)
2) land_type=Tree 2868 282 Yes (0.90167364 0.09832636) *
3) land_type=Barren,Non-tree vegetation 2462 346 No (0.14053615 0.85946385)
6) temp_annual_max< 13.385 323 143 Yes (0.55727554 0.44272446)
12) tree_no_tree=Tree 84 6 Yes (0.92857143 0.07142857) *
13) tree_no_tree=No tree 239 102 No (0.42677824 0.57322176) *
7) temp_annual_max>=13.385 2139 166 No (0.07760636 0.92239364) *
Fit with a workflow:
workflow ( ) |> add_formula ( forested ~ . ) |>
add_model ( tree_spec ) |>
fit ( data = forested_train )
══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Formula
Model: decision_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
forested ~ .
── Model ───────────────────────────────────────────────────────────────────────
n= 5330
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5330 2398 Yes (0.55009381 0.44990619)
2) land_type=Tree 2868 282 Yes (0.90167364 0.09832636) *
3) land_type=Barren,Non-tree vegetation 2462 346 No (0.14053615 0.85946385)
6) temp_annual_max< 13.385 323 143 Yes (0.55727554 0.44272446)
12) tree_no_tree=Tree 84 6 Yes (0.92857143 0.07142857) *
13) tree_no_tree=No tree 239 102 No (0.42677824 0.57322176) *
7) temp_annual_max>=13.385 2139 166 No (0.07760636 0.92239364) *
“Shortcut” by specifying the preprocessor and model spec directly in the workflow() call:
workflow ( forested ~ . , tree_spec ) |> fit ( data = forested_train )
══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Formula
Model: decision_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
forested ~ .
── Model ───────────────────────────────────────────────────────────────────────
n= 5330
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 5330 2398 Yes (0.55009381 0.44990619)
2) land_type=Tree 2868 282 Yes (0.90167364 0.09832636) *
3) land_type=Barren,Non-tree vegetation 2462 346 No (0.14053615 0.85946385)
6) temp_annual_max< 13.385 323 143 Yes (0.55727554 0.44272446)
12) tree_no_tree=Tree 84 6 Yes (0.92857143 0.07142857) *
13) tree_no_tree=No tree 239 102 No (0.42677824 0.57322176) *
7) temp_annual_max>=13.385 2139 166 No (0.07760636 0.92239364) *
Your turn
Edit the chunk below to make a workflow with your own model of choice!
Extension/Challenge: Other than formulas, what kinds of preprocessors are supported?
tree_spec <- decision_tree ( ) |> set_mode ( "classification" )
tree_wflow <- workflow ( ) |> add_formula ( forested ~ . ) |>
add_model ( tree_spec )
tree_wflow
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Formula
Model: decision_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
forested ~ .
── Model ───────────────────────────────────────────────────────────────────────
Decision Tree Model Specification (classification)
Computational engine: rpart
log_spec <- logistic_reg ( ) |> set_mode ( "classification" )
log_wflow <- workflow ( ) |> add_formula ( forested ~ . ) |>
add_model ( log_spec )
log_wflow
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Formula
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
forested ~ .
── Model ───────────────────────────────────────────────────────────────────────
Logistic Regression Model Specification (classification)
Computational engine: glm
Predict with your model
tree_fit <- workflow ( forested ~ . , tree_spec ) |> fit ( data = forested_train )
Your turn
Run:
predict ( tree_fit , new_data = forested_test )
# A tibble: 1,777 × 1
.pred_class
<fct>
1 Yes
2 Yes
3 No
4 No
5 No
6 Yes
7 Yes
8 Yes
9 Yes
10 Yes
# ℹ 1,767 more rows
What do you notice about the structure of the result?
Add response here. It only contains the predicted class value (Yes/No). No probability values, no predictors, residuals, etc.
Your turn
Run:
augment ( tree_fit , new_data = forested_test )
# A tibble: 1,777 × 22
.pred_class .pred_Yes .pred_No forested year elevation eastness northness
<fct> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>
1 Yes 0.902 0.0983 No 2005 164 -84 53
2 Yes 0.902 0.0983 Yes 2005 299 93 34
3 No 0.0776 0.922 Yes 2005 636 -48 87
4 No 0.0776 0.922 Yes 2005 224 -65 -75
5 No 0.427 0.573 Yes 2005 2240 -67 -74
6 Yes 0.902 0.0983 Yes 2004 1044 96 -26
7 Yes 0.902 0.0983 Yes 2003 1031 -49 86
8 Yes 0.902 0.0983 Yes 2005 1330 99 7
9 Yes 0.902 0.0983 Yes 2005 1423 46 88
10 Yes 0.902 0.0983 Yes 2014 546 -92 -38
# ℹ 1,767 more rows
# ℹ 14 more variables: roughness <dbl>, tree_no_tree <fct>, dew_temp <dbl>,
# precip_annual <dbl>, temp_annual_mean <dbl>, temp_annual_min <dbl>,
# temp_annual_max <dbl>, temp_january_min <dbl>, vapor_min <dbl>,
# vapor_max <dbl>, canopy_cover <dbl>, lon <dbl>, lat <dbl>, land_type <fct>
How does the output compare to the output from predict()
Add response here. Has the predicted class value, probabilities, actual class value, and predictors.
Understand your model
Loading required package: rpart
Attaching package: 'rpart'
The following object is masked from 'package:dials':
prune
tree_fit |> extract_fit_engine ( ) |>
rpart.plot ( roundint = FALSE )
Your turn
Try extracting the model engine object from your fitted workflow!
# add code here log_wflow |> fit ( data = forested_train ) |>
extract_fit_engine ( )
Call: stats::glm(formula = ..y ~ ., family = stats::binomial, data = data)
Coefficients:
(Intercept) year
-9.5066243 0.0025324
elevation eastness
-0.0021492 -0.0005322
northness roughness
0.0025026 -0.0037322
`tree_no_treeNo tree` dew_temp
1.3100053 -0.1688934
precip_annual temp_annual_mean
-0.0001162 -6.4815711
temp_annual_min temp_annual_max
0.7398032 2.2873451
temp_january_min vapor_min
3.1113837 -0.0021930
vapor_max canopy_cover
0.0088182 -0.0468181
lon lat
-0.0876664 0.0919399
`land_typeNon-tree vegetation` land_typeTree
-0.8998639 -1.6773218
Degrees of Freedom: 5329 Total (i.e. Null); 5310 Residual
Null Deviance: 7335
Residual Deviance: 2609 AIC: 2649
What kind of object is it? What can you do with it?
Add response here It’s a glm object. You can use it to extract coefficients, residuals, etc., using base R methods.
Acknowledgments
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.1 (2024-06-14)
os macOS Sonoma 14.6.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-09-09
pandoc 3.3 @ /usr/local/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
backports 1.5.0 2024-05-23 [1] CRAN (R 4.4.0)
broom * 1.0.6 2024-05-17 [1] CRAN (R 4.4.0)
class 7.3-22 2023-05-03 [1] CRAN (R 4.4.0)
cli 3.6.3 2024-06-21 [1] CRAN (R 4.4.0)
codetools 0.2-20 2024-03-31 [1] CRAN (R 4.4.1)
colorspace 2.1-1 2024-07-26 [1] CRAN (R 4.4.0)
data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.1)
dials * 1.3.0 2024-07-30 [1] CRAN (R 4.4.0)
DiceDesign 1.10 2023-12-07 [1] CRAN (R 4.3.1)
digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
evaluate 0.24.0 2024-06-10 [1] CRAN (R 4.4.0)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.2 2024-05-13 [1] CRAN (R 4.3.3)
fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
forested * 0.1.0 2024-07-31 [1] CRAN (R 4.4.0)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.2 2024-03-26 [1] CRAN (R 4.3.1)
future.apply 1.11.2 2024-03-28 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.3.1)
globals 0.16.3 2024-03-08 [1] CRAN (R 4.3.1)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gtable 0.3.5 2024-04-22 [1] CRAN (R 4.3.1)
hardhat 1.4.0 2024-06-02 [1] CRAN (R 4.4.0)
here 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
infer * 1.0.7 2024-03-25 [1] CRAN (R 4.3.1)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
knitr 1.47 2024-05-29 [1] CRAN (R 4.4.0)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
lattice 0.22-6 2024-03-20 [1] CRAN (R 4.4.0)
lava 1.8.0 2024-03-05 [1] CRAN (R 4.3.1)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.1)
lubridate 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-61 2024-06-13 [1] CRAN (R 4.4.0)
Matrix 1.7-0 2024-03-22 [1] CRAN (R 4.4.0)
modeldata * 1.4.0 2024-06-19 [1] CRAN (R 4.4.0)
modelenv 0.1.1 2023-03-08 [1] CRAN (R 4.3.0)
munsell 0.5.1 2024-04-01 [1] CRAN (R 4.3.1)
nnet 7.3-19 2023-05-03 [1] CRAN (R 4.4.0)
parallelly 1.37.1 2024-02-29 [1] CRAN (R 4.3.1)
parsnip * 1.2.1 2024-03-22 [1] CRAN (R 4.3.1)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
prodlim 2023.08.28 2023-08-28 [1] CRAN (R 4.3.0)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
recipes * 1.0.10 2024-02-18 [1] CRAN (R 4.3.1)
rlang 1.1.4 2024-06-04 [1] CRAN (R 4.3.3)
rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
rpart * 4.1.23 2023-12-05 [1] CRAN (R 4.4.0)
rpart.plot * 3.1.2 2024-02-26 [1] CRAN (R 4.4.0)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
rsample * 1.2.1 2024-03-25 [1] CRAN (R 4.3.1)
rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.3.1)
scales * 1.3.0.9000 2024-05-07 [1] Github (r-lib/scales@c0f79d3)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
survival 3.7-0 2024-06-05 [1] CRAN (R 4.4.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidymodels * 1.2.0 2024-03-25 [1] CRAN (R 4.3.1)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.1)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
timeDate 4032.109 2023-12-14 [1] CRAN (R 4.3.1)
tune * 1.2.1 2024-04-18 [1] CRAN (R 4.3.1)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
withr 3.0.1 2024-07-31 [1] CRAN (R 4.4.0)
workflows * 1.1.4 2024-02-19 [1] CRAN (R 4.3.1)
workflowsets * 1.1.0 2024-03-21 [1] CRAN (R 4.3.1)
xfun 0.45 2024-06-16 [1] CRAN (R 4.4.0)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
yardstick * 1.3.1 2024-03-21 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────