library(tidyverse)
library(tidymodels)
library(tidytext)
library(embed)
library(colorspace)
set.seed(5312)
# preferred theme
theme_set(theme_minimal(base_size = 12, base_family = "Atkinson Hyperlegible"))AE 10: Dimension reduction with cocktails
Suggested answers
Cocktails
Tidy Tuesday published a data set on cocktail recipes. In this application exercise, we will use dimension reduction techniques to explore the data set based on the recipes’ ingredients.
cocktails <- read_csv("data/boston_cocktails.csv")
cocktails# A tibble: 3,643 × 6
name category row_id ingredient_number ingredient measure
<chr> <chr> <dbl> <dbl> <chr> <chr>
1 Gauguin Cocktail Classics 1 1 Light Rum 2 oz
2 Gauguin Cocktail Classics 1 2 Passion F… 1 oz
3 Gauguin Cocktail Classics 1 3 Lemon Jui… 1 oz
4 Gauguin Cocktail Classics 1 4 Lime Juice 1 oz
5 Fort Lauderdale Cocktail Classics 2 1 Light Rum 1 1/2 …
6 Fort Lauderdale Cocktail Classics 2 2 Sweet Ver… 1/2 oz
7 Fort Lauderdale Cocktail Classics 2 3 Juice of … 1/4 oz
8 Fort Lauderdale Cocktail Classics 2 4 Juice of … 1/4 oz
9 Apple Pie Cordials and Liq… 3 1 Apple sch… 3 oz
10 Apple Pie Cordials and Liq… 3 2 Cinnamon … 1 oz
# ℹ 3,633 more rowsData cleaning
cocktails_parsed <- cocktails |>
mutate(
ingredient = str_to_lower(ingredient),
ingredient = str_replace_all(ingredient, "-", " "),
ingredient = str_remove(ingredient, " liqueur$"),
ingredient = str_remove(ingredient, " (if desired)$"),
ingredient = case_when(
str_detect(ingredient, "bitters") ~ "bitters",
str_detect(ingredient, "lemon") ~ "lemon juice",
str_detect(ingredient, "lime") ~ "lime juice",
str_detect(ingredient, "grapefruit") ~ "grapefruit juice",
str_detect(ingredient, "orange") ~ "orange juice",
.default = ingredient
),
measure = case_when(
str_detect(ingredient, "bitters") ~ str_replace(measure, "oz$", "dash"),
.default = measure
),
measure = str_replace(measure, " ?1/2", ".5"),
measure = str_replace(measure, " ?3/4", ".75"),
measure = str_replace(measure, " ?1/4", ".25"),
measure_number = parse_number(measure),
measure_number = if_else(str_detect(measure, "dash$"),
measure_number / 50,
measure_number
)
) |>
# only keep ingredients that appear more than 15 times
filter(n() >= 15, .by = ingredient) |>
distinct(row_id, ingredient, .keep_all = TRUE) |>
drop_na()
cocktails_parsed# A tibble: 2,602 × 7
name category row_id ingredient_number ingredient measure measure_number
<chr> <chr> <dbl> <dbl> <chr> <chr> <dbl>
1 Gauguin Cocktai… 1 1 light rum 2 oz 2
2 Gauguin Cocktai… 1 3 lemon jui… 1 oz 1
3 Gauguin Cocktai… 1 4 lime juice 1 oz 1
4 Fort Lau… Cocktai… 2 1 light rum 1.5 oz 1.5
5 Fort Lau… Cocktai… 2 2 sweet ver… .5 oz 0.5
6 Fort Lau… Cocktai… 2 3 orange ju… .25 oz 0.25
7 Fort Lau… Cocktai… 2 4 lime juice .25 oz 0.25
8 Cuban Co… Cocktai… 4 1 lime juice .5 oz 0.5
9 Cuban Co… Cocktai… 4 2 powdered … .5 oz 0.5
10 Cuban Co… Cocktai… 4 3 light rum 2 oz 2
# ℹ 2,592 more rows# convert to wide format for modeling
cocktails_df <- cocktails_parsed |>
select(-ingredient_number, -row_id, -measure) |>
pivot_wider(names_from = ingredient, values_from = measure_number, values_fill = 0) |>
janitor::clean_names() |>
drop_na()
cocktails_df# A tibble: 943 × 46
name category light_rum lemon_juice lime_juice sweet_vermouth orange_juice
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Gauguin Cocktai… 2 1 1 0 0
2 Fort L… Cocktai… 1.5 0 0.25 0.5 0.25
3 Cuban … Cocktai… 2 0 0.5 0 0
4 Cool C… Cocktai… 0 0 0 0 1
5 John C… Whiskies 0 1 0 0 0
6 Cherry… Cocktai… 1.25 0 0 0 0
7 Casa B… Cocktai… 2 0 1.5 0 0
8 Caribb… Cocktai… 0.5 0 0 0 0
9 Amber … Cordial… 0 0.25 0 0 0
10 The Jo… Whiskies 0 0.5 0 0 0
# ℹ 933 more rows
# ℹ 39 more variables: powdered_sugar <dbl>, dark_rum <dbl>,
# cranberry_juice <dbl>, pineapple_juice <dbl>, bourbon_whiskey <dbl>,
# simple_syrup <dbl>, cherry_flavored_brandy <dbl>, light_cream <dbl>,
# triple_sec <dbl>, maraschino <dbl>, amaretto <dbl>, grenadine <dbl>,
# apple_brandy <dbl>, brandy <dbl>, gin <dbl>, anisette <dbl>,
# dry_vermouth <dbl>, apricot_flavored_brandy <dbl>, sloe_gin <dbl>, …Principal components analysis
Estimate PCA
Your turn: Apply PCA to the data set. Define and prepare a feature engineering recipe that scales the ingredients features and then applies PCA.
pca_rec <- recipe(~., data = cocktails_df) |>
update_role(name, category, new_role = "id") |>
step_normalize(all_predictors()) |>
step_pca(all_predictors())
pca_prep <- prep(pca_rec)
pca_prepInterpret results
Your turn: Explore the results of the PCA by identifying how each ingredient loads (contributes to) the first five principal components. How would you describe each component? What do they seem to measure?
tidied_pca <- tidy(pca_prep, 2)
tidied_pca |>
filter(component %in% str_glue("PC{1:5}")) |>
mutate(component = fct_inorder(component)) |>
ggplot(mapping = aes(value, terms, fill = terms)) +
geom_col(show.legend = FALSE) +
scale_color_discrete_qualitative() +
facet_wrap(facets = vars(component), nrow = 1) +
labs(y = NULL)Add response here. PC1 seems to indicate the difference between sweeteners used in cocktails (e.g. powdered sugar vs. simple syrup). PC2 seems defined by the contribution of vermouths. It gets harder to interpret the others, especially given the number of unique ingredients.
tidied_pca |>
# only first four components
filter(component %in% str_glue("PC{1:4}")) |>
# top 8 ingredients per component
slice_max(order_by = abs(value), n = 10, by = component) |>
# reorder the terms in decreasing magnitude for visualization
mutate(terms = reorder_within(terms, abs(value), component)) |>
ggplot(mapping = aes(x = value, y = terms, fill = value > 0)) +
geom_col(show.legend = FALSE) +
facet_wrap(facets = vars(component), scales = "free_y") +
scale_y_reordered() +
labs(
x = "Value of contribution",
y = NULL
)Add response here. The top ingredients contributing to PC1 are sweeteners (powdered sugar, simple syrup) and ingredients for a margaritas (simple syrup, lime juice, tequila). PC2 is defined by vermouths (sweet, dry). PC3 is more difficult to interpret, but it seems to be defined by the contribution of egg whites and grenadine (amaretto sours and their variants?). PC4 is defined by the contribution of vodka and OJ (screwdrivers?). In the opposite direction are ingredients for rum-based drinks (light rum, triple sec, lime juice, simple syrup).
Your turn: How are the beverages distributed along the first two dimensions? Is there a pattern to these dimensions?
cocktails_pca <- bake(pca_prep, new_data = NULL) |>
# collapse infrequent categories
mutate(category = fct_lump_lowfreq(f = category))
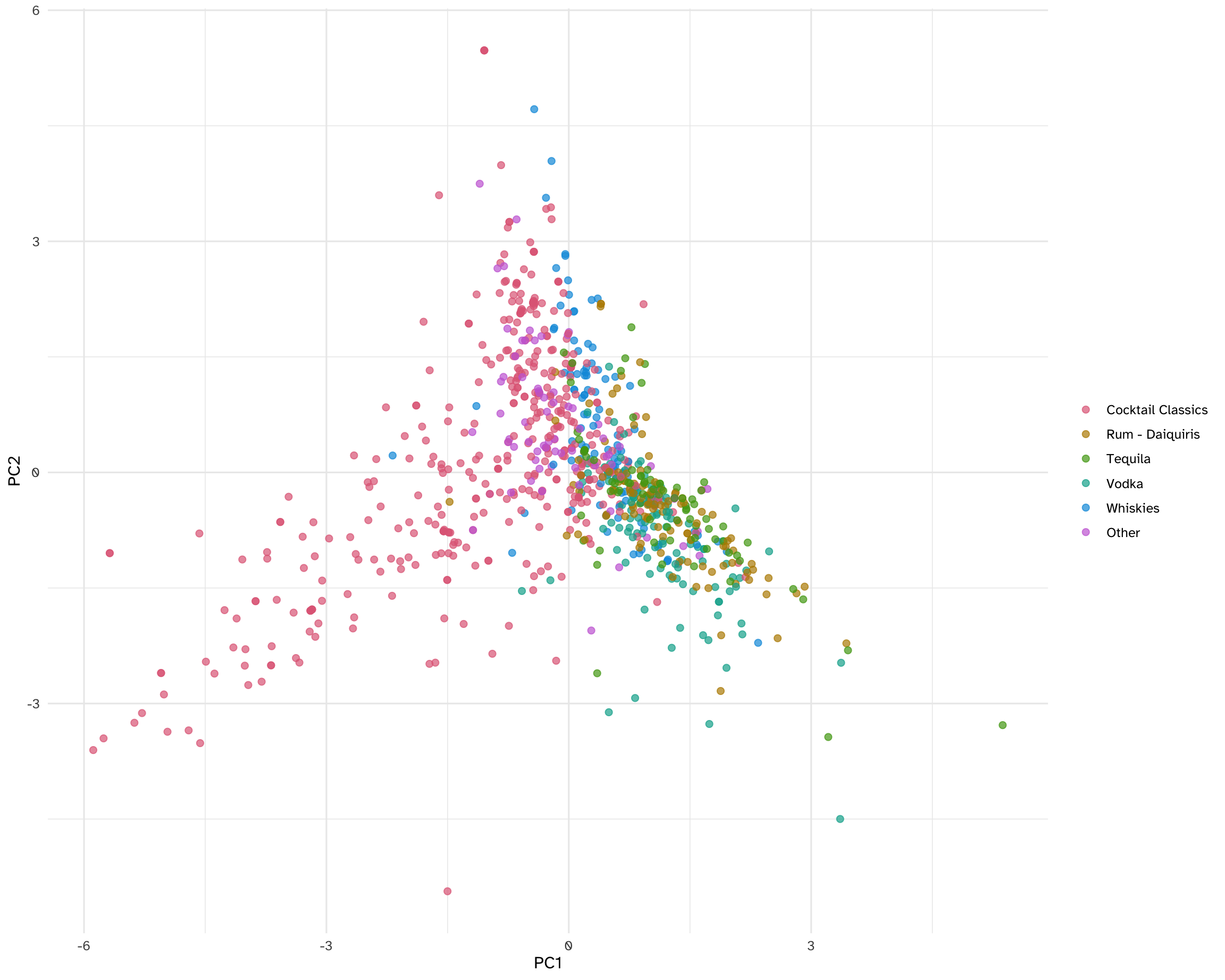
ggplot(data = cocktails_pca, mapping = aes(x = PC1, y = PC2, label = name)) +
geom_point(mapping = aes(color = category), alpha = 0.7, size = 2) +
geom_text(check_overlap = TRUE, hjust = "inward", family = "Atkinson Hyperlegible") +
scale_color_discrete_qualitative() +
labs(color = NULL)ggplot(data = cocktails_pca, mapping = aes(x = PC1, y = PC2, label = name)) +
geom_point(mapping = aes(color = category), alpha = 0.7, size = 2) +
scale_color_discrete_qualitative() +
labs(color = NULL)# first four components
library(GGally)
ggpairs(
data = cocktails_pca,
columns = 3:6,
lower = list(
mapping = aes(color = category)
),
diag = list(
mapping = aes(color = category, fill = category)
),
progress = FALSE
) +
scale_color_discrete_qualitative() +
scale_fill_discrete_qualitative() +
labs(
color = "Label",
fill = "Label"
)Add response here. PC1 has mostly “Cocktail classics” on the left and tequila/vodka on the right. Whiskies and “other” are in the middle.
Your turn: How much do the dimensions explain variation in the data set? If you were going to use these components for a predictive model, how many would you include?
tidy(pca_prep, number = 2, type = "variance") |>
filter(terms == "percent variance") |>
ggplot(mapping = aes(x = component, y = value)) +
geom_line() +
scale_y_continuous(labels = label_percent(scale = 1)) +
labs(
x = "Principal component",
y = "Percent variance"
)Add response here. PCA is kind of doing poorly here. The first components by definition have the largest amount of variance, but overall is exceedingly small. I would probably include the first 5 components definitely, if not more. Or frankly, depending on the model I might just keep the original ingredient measures.
UMAP
Your turn: Apply UMAP to the data set. Define and prepare a feature engineering recipe that scales the ingredients features and then applies UMAP, retaining four UMAP dimensions. Examine the location of cocktails along the first two UMAP dimensions. How does this compare to the PCA results?
set.seed(532)
umap_rec <- recipe(~., data = cocktails_df) |>
update_role(name, category, new_role = "id") |>
step_normalize(all_predictors()) |>
step_umap(all_predictors(), num_comp = 4)
umap_prep <- prep(umap_rec)
umap_prepbake(umap_prep, new_data = NULL) |>
# collapse infrequent categories
mutate(category = fct_lump_lowfreq(f = category)) |>
ggplot(mapping = aes(x = UMAP1, y = UMAP2, label = name)) +
geom_point(mapping = aes(color = category), alpha = 0.7, size = 2) +
geom_text(check_overlap = TRUE, hjust = "inward", family = "Atkinson Hyperlegible") +
scale_color_discrete_qualitative() +
labs(color = NULL)bake(umap_prep, new_data = NULL) |>
# collapse infrequent categories
mutate(category = fct_lump_lowfreq(f = category)) |>
ggplot(mapping = aes(x = UMAP1, y = UMAP2, label = name)) +
geom_point(mapping = aes(color = category), alpha = 0.7, size = 2) +
scale_color_discrete_qualitative() +
labs(color = NULL)# first four dimensions
umap_prep |>
bake(new_data = NULL) |>
# collapse infrequent categories
mutate(category = fct_lump_lowfreq(f = category)) |>
ggpairs(
columns = 3:6,
lower = list(
mapping = aes(color = category)
),
diag = list(
mapping = aes(color = category, fill = category)
),
progress = FALSE
) +
scale_color_discrete_qualitative() +
scale_fill_discrete_qualitative() +
labs(
color = "Label",
fill = "Label"
)Add response here. Clearly a more localized structure to the beverages, but not discernable trends globally.
Acknowledgments
- Application exercise drawn from PCA and UMAP with tidymodels and #TidyTuesday cocktail recipes by Julia Silge and licensed under CC BY-SA 4.0.
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.1 (2024-06-14)
os macOS Sonoma 14.6.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-10-03
pandoc 3.4 @ /usr/local/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
archive 1.1.8 2024-04-28 [1] CRAN (R 4.4.0)
backports 1.5.0 2024-05-23 [1] CRAN (R 4.4.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
broom * 1.0.6 2024-05-17 [1] CRAN (R 4.4.0)
class 7.3-22 2023-05-03 [1] CRAN (R 4.4.0)
cli 3.6.3 2024-06-21 [1] CRAN (R 4.4.0)
codetools 0.2-20 2024-03-31 [1] CRAN (R 4.4.1)
colorspace * 2.1-1 2024-07-26 [1] CRAN (R 4.4.0)
crayon 1.5.3 2024-06-20 [1] CRAN (R 4.4.0)
data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.1)
dials * 1.3.0 2024-07-30 [1] CRAN (R 4.4.0)
DiceDesign 1.10 2023-12-07 [1] CRAN (R 4.3.1)
digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
embed * 1.1.4 2024-03-20 [1] CRAN (R 4.3.1)
evaluate 0.24.0 2024-06-10 [1] CRAN (R 4.4.0)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.2 2024-05-13 [1] CRAN (R 4.3.3)
fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.2 2024-03-26 [1] CRAN (R 4.3.1)
future.apply 1.11.2 2024-03-28 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
GGally * 2.2.1 2024-02-14 [1] CRAN (R 4.4.0)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.3.1)
ggstats 0.6.0 2024-04-05 [1] CRAN (R 4.4.0)
globals 0.16.3 2024-03-08 [1] CRAN (R 4.3.1)
glue 1.8.0 2024-09-30 [1] CRAN (R 4.4.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gtable 0.3.5 2024-04-22 [1] CRAN (R 4.3.1)
hardhat 1.4.0 2024-06-02 [1] CRAN (R 4.4.0)
here 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
infer * 1.0.7 2024-03-25 [1] CRAN (R 4.3.1)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.4.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
janeaustenr 1.0.0 2022-08-26 [1] CRAN (R 4.3.0)
janitor 2.2.0 2023-02-02 [1] CRAN (R 4.3.0)
jsonlite 1.8.9 2024-09-20 [1] CRAN (R 4.4.1)
knitr 1.47 2024-05-29 [1] CRAN (R 4.4.0)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
lattice 0.22-6 2024-03-20 [1] CRAN (R 4.4.0)
lava 1.8.0 2024-03-05 [1] CRAN (R 4.3.1)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.1)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-61 2024-06-13 [1] CRAN (R 4.4.0)
Matrix 1.7-0 2024-03-22 [1] CRAN (R 4.4.0)
modeldata * 1.4.0 2024-06-19 [1] CRAN (R 4.4.0)
munsell 0.5.1 2024-04-01 [1] CRAN (R 4.3.1)
nnet 7.3-19 2023-05-03 [1] CRAN (R 4.4.0)
parallelly 1.37.1 2024-02-29 [1] CRAN (R 4.3.1)
parsnip * 1.2.1 2024-03-22 [1] CRAN (R 4.3.1)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.1)
prodlim 2023.08.28 2023-08-28 [1] CRAN (R 4.3.0)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
Rcpp 1.0.13 2024-07-17 [1] CRAN (R 4.4.0)
RcppAnnoy 0.0.22 2024-01-23 [1] CRAN (R 4.3.1)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
recipes * 1.0.10 2024-02-18 [1] CRAN (R 4.3.1)
rlang 1.1.4 2024-06-04 [1] CRAN (R 4.3.3)
rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
rpart 4.1.23 2023-12-05 [1] CRAN (R 4.4.0)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
rsample * 1.2.1 2024-03-25 [1] CRAN (R 4.3.1)
rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.3.1)
scales * 1.3.0 2023-11-28 [1] CRAN (R 4.4.0)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
snakecase 0.11.1 2023-08-27 [1] CRAN (R 4.3.0)
SnowballC 0.7.1 2023-04-25 [1] CRAN (R 4.3.0)
stringi 1.8.4 2024-05-06 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
survival 3.7-0 2024-06-05 [1] CRAN (R 4.4.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidymodels * 1.2.0 2024-03-25 [1] CRAN (R 4.3.1)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.1)
tidytext * 0.4.2 2024-04-10 [1] CRAN (R 4.4.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
timeDate 4032.109 2023-12-14 [1] CRAN (R 4.3.1)
tokenizers 0.3.0 2022-12-22 [1] CRAN (R 4.3.0)
tune * 1.2.1 2024-04-18 [1] CRAN (R 4.3.1)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
uwot 0.2.2 2024-04-21 [1] CRAN (R 4.4.0)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vroom 1.6.5 2023-12-05 [1] CRAN (R 4.3.1)
withr 3.0.1 2024-07-31 [1] CRAN (R 4.4.0)
workflows * 1.1.4 2024-02-19 [1] CRAN (R 4.3.1)
workflowsets * 1.1.0 2024-03-21 [1] CRAN (R 4.3.1)
xfun 0.45 2024-06-16 [1] CRAN (R 4.4.0)
yaml 2.3.10 2024-07-26 [1] CRAN (R 4.4.0)
yardstick * 1.3.1 2024-03-21 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────